Abstract
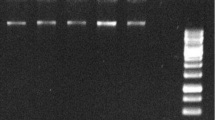
Isolation of high-quality RNA from Dendrobium flowers is challenging because of the high levels of pigment, polysaccharides, and polyphenols. In the present study, an efficient CTAB method for RNA extraction from the pigment-rich flowers of Dendrobium was optimised. The optimised method yielded high quantities of RNA (10.1–12.9 µg/g). Spectrophotometric values of A260/280 in the range of 2.2 to 2.4 and A260/230 values of 2.0 suggested that the isolated RNA was free of polyphenols, polysaccharides, and protein contaminants. RNA integrity numbers determined by microfluidics were in the range of 7.9–8.9 indicative of intact RNA. In the improved method, the addition of 3 M NaCl and 3% PVP-10 in the extraction buffer, followed by an incubation period of 45 min at 65 °C, eliminated most of the polysaccharides, polyphenolic compounds, and denatured protein. Extraction with phenol:chloroform:isoamyl alcohol (125:24:1) effectively removed pigments from the aqueous phase, while the precipitation of RNA with lithium chloride minimised the co-precipitation of protein, DNA, and polysaccharide and resulted in the extraction of high quality of RNA. The suitability of the RNA for downstream processing was confirmed via RT-PCR amplification of Chalcone synthase gene from cDNA prepared from RNA isolated from different developmental stages of the flower of a Dendrobium hybrid. The present method will be highly useful for the isolation of RNA from pigment, polyphenol, and polysaccharide-rich plant tissues.






Similar content being viewed by others
Abbreviations
- bp:
-
Base pair
- cDNA:
-
Complimentary DNA
- CHS:
-
Chalcone synthase
- C:I:
-
Chloroform:isoamyl alcohol
- P:C:I:
-
Phenol:chloroform:isoamyl alcohol
- CTAB:
-
Cetyltrimethylammonium bromide
- °C:
-
Degrees Celsius
- cm:
-
Centimetre
- DEPC:
-
Diethyl pyrocarbonate
- DNase:
-
Deoxyribonuclease
- EDTA:
-
Ethylenediaminetetraacetic acid
- EtBr:
-
Ethidium bromide
- FW:
-
Fresh weight
- LiCl:
-
Lithium chloride
- M:
-
Molarity
- ml:
-
Millilitre
- mM:
-
Millimolar
- min:
-
Minutes
- NaCl:
-
Sodium chloride
- psi:
-
Pound-force per square inch
- PVP-10:
-
Polyvinylpyrrolidone-10
- RNase:
-
Ribonuclease
- rRNA:
-
Ribosomal RNA
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- s:
-
Second
- SDS:
-
Sodium dodecyl sulfate
- TBE:
-
Tris-borate-EDTA
- Tris–HCl:
-
Tris hydrochloride
- µl:
-
Microlitre
- ng:
-
Nanogram
- w/v:
-
Weight per volume
- v/v:
-
Volume per volume
- %:
-
Percentage
References
Asif M, Trivedi P, Solomos T, Tucker M (2006) Isolation of high-quality RNA from apple (Malus domestica) fruit. J Agric Food Chem 54:5227–5229. https://doi.org/10.1021/jf053137n
Barman P, Choudhary AK, Geeta R (2017) A modified protocol yields high-quality RNA from highly mucilaginous Dioscorea tubers. 3 Biotech 7:150. https://doi.org/10.1007/s13205-017-0775-9
Carpenter A, Siggia S, Carter S (1976) Separation and/or concentration of phenolic materials from dilute aqueous solutions. Anal Chem 48:225–228
Choi C, Yoon S, Moon H, Bae YU, Kim CB, Diskul-Na-Ayudthaya P, Ngu TV, Munir J, Han J, Park SB, Moon JS (2018) mirRICH, a simple method to enrich the small RNA fraction from over-dried RNA pellets. RNA Biol 28:1
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
De LC, Medhi RP (2015) Orchid–a diversified component of farming systems for profitability and livelihood security of small and marginal farmers. J Glob Biosci 4:1393–1406
De LC, Pathak P, Rao AN, Rajeevan PK (2014) Commercial orchids National Research Centre for Orchids, India
Deepa K, Sheeja TE, Santhi R, Sasikumar B, Cyriac A, Deepesh PV, Prasath D (2014) A simple and efficient protocol for isolation of high quality functional RNA from different tissues of turmeric (Curcuma longa L.). Physiol Mol Biol Plants 20:263–267. https://doi.org/10.1007/s12298-013-0218-y
El-Ashram S, Al Nasr I, Suo X (2016) Nucleic acid protocols: extraction and optimization. Biotechnol Rep 12:33–39. https://doi.org/10.1016/j.btre.2016.10.001
Fang G, Hammar S, Grumet R (1992) A quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biotechniques 13(52–54):56
Gao Y, Guangqi Z, Jiang C, Yao S, Kang YE, Shucheng F (2016) Comparison of Different Methods for RNA Extraction from Floral Buds of Tree Peony (Paeonia suffruticosa Andr.). Notulae Botanicae Horti Agrobotanici Cluj-Napoca 44:418–422
Govaerts R (2003) World checklist of monocotyledons database in ACCESS: 1-71827 The Board of Trustees of the Royal Botanic Gardens, Kew
Harisha BN (2017) An economic analysis of floriculture in India. Int J Acad Res Dev. https://doi.org/10.22271/academic
Hinsley A, de Boer HJ, Fay MF, Gale SW, Gardiner LM, Gunasekara RS, Kumar P, Masters S, Metusala D, Roberts DL, Veldman S (2017) A review of the trade in orchids and its implications for conservation. Bot J Linn Soc 186:435–455
Huded AK, Jingade P, Mishra MK (2018) A rapid and efficient SDS-based RNA isolation protocol from different tissues of coffee. 3 Biotech 8:183
Kiefer E, Heller W, Ernst D (2000) A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol Biol Report 18:33–39. https://doi.org/10.1007/BF02825291
Lau S-E, Schwarzacher T, Othman RY, Harikrishna JA (2015) dsRNA silencing of an R2R3-MYB transcription factor affects flower cell shape in a Dendrobium hybrid. BMC Plant Biol 15:194. https://doi.org/10.1186/s12870-015-0577-3
Lee WS, Gudimella R, Wong GR, Tammi MT, Khalid N (2015) Harikrishna JA (2015) Transcripts and microRNAs responding to salt stress in Musa acuminata Colla (AAA Group) cv Berangan roots. PLoS One 10(5):e0127526
Leh TY, Yong CS, Nulit R, Abdullah JO (2019) Efficient and high-quality RNA isolation from metabolite-rich tissues of Stevia rebaudiana, an important commercial crop. Trop Life Sci Res 30:149
Lehninger AL, Nelson DL, Cox MM (2005) Lehninger principles of biochemistry. WH Freeman, New York
Li R (2015) Forensic biology. CRC Press, Boca Raton
Liu L, Han R, Yu N, Zhang W, Xing L, Xie D, Peng D (2018) A method for extracting high-quality total RNA from plant rich in polysaccharides and polyphenols using Dendrobium huoshanense. PLOS One 13:e0196592. https://doi.org/10.1371/journal.pone.0196592
Moretti M, Cossignani L, Messina F, Dominici L, Villarini M, Curini M, Marcotullio MC (2013) Antigenotoxic effect, composition and antioxidant activity of Dendrobium speciosum. Food Chem 140:660–665. https://doi.org/10.1016/j.foodchem.2012.10.022
Muoki RC, Paul A, Kumari A, Singh K, Kumar S (2012) An improved protocol for the isolation of RNA from roots of tea (Camellia sinensis (L.) O. Kuntze). Mol Biotechnol 52:82–88. https://doi.org/10.1007/s12033-011-9476-5
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Qadri R, Iqbal A, Wu Y, Li J, Nisar N, Azam M, Yang Y (2019) A modified protocol for total RNA isolation from different oil palm (Elaeis guineensis) tissues using cetyltrimethylammonium bromide. Curr Sci 116:479–482
Raizada A, Jegadeesan S (2019) An improved method for rapid isolation of DNA and RNA from leaves, flowers and roots of blackgram [Vigna mungo (L.) Hepper] for detection of begomovirus infection and RT-PCR. Electron J Plant Breed 10:167–176
Rodríguez-Ávila NL, Narváez-Zapata JA, Aguilar-Espinosa M, Rivera-Madrid R (2011) Regulation of pigment-related genes during flower and fruit development of Bixa orellana. Plant Mol Biol Report 29:43–50
Sarmah D, Kolukunde S, Sutradhar M, Singh BK, Mandal T, Mandal N (2017) A review on: in vitro cloning of orchids. Int J Curr Microbiol Appl Sci 6:1909–1927
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 1:3. https://doi.org/10.1186/1471-2199-7-3
Shu C, Sun S, Chen J, Chen J, Zhou E (2014) Comparison of different methods for total RNA extraction from sclerotia of Rhizoctonia solani. Electron J Biotechnol 17:50–54
Su CL, Chao YT, Alex Chang YC, Chen WC, Chen CY, Lee AY, Hwa KT, Shih MC (2011) De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol 52:1501–1514. https://doi.org/10.1093/pcp/pcr097
Surzycki S (2012) Basic techniques in molecular biology. Springer Science & Business Media, Berlin
The nation Thailand (2018) https://www.nationthailand.com/breakingnews/30360623. Accessed 5 Sept 2019
Toni LS, Garcia AM, Jeffrey DA, Jiang X, Stauffer BL, Miyamoto SD, Sucharov CC (2018) Optimization of phenol-chloroform RNA extraction. MethodsX 5:599–608
Wong L-M, Silvaraj S, Phoon L-Q (2014) An optimised high-salt CTAB protocol for both DNA and RNA isolation from succulent stems of Hylocereus sp. J Med Bioeng 3:236–240
Acknowledgements
This work was supported by the Frontier Research Grant (FRG 2017, FG026-17AFR) and CEBAR Research University Grants (RU006-2017 and RU006-2018).
Author information
Authors and Affiliations
Contributions
MAKA, PM, S-EL, and TTT conducted experimental work; PM and JAH made the experimental design. All authors contributed to manuscript preparation and to editing of both original and revised manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13205_2019_1898_MOESM2_ESM.pptx
Supplementary Fig. S1 Flow diagram of cDNA synthesis from total RNA (SuperScript® III First-Strand Synthesis system (Invitrogen, USA)
13205_2019_1898_MOESM3_ESM.pptx
Supplementary Fig. S2 Chalcone synthase (CHS) gene sequence of Dendrobium hybrid cultivar Sonia Earsakul (KC345011.1) with primer binding sites (Orange letter denotes introns and blue- bold letter denotes primer binding sites)
13205_2019_1898_MOESM4_ESM.pptx
Supplementary Fig. S3 Aqueous fraction collected after P:C:I steps of RNA isolation method. M1: RNA isolation method of Kiefer et al. 2000, M2: RNA isolation method of Su et al. 2011, M3: RNA isolation method of Gao et al. 2016, M4: RNA isolation method of Liu et al. 2018, and M5: improved CTAB method
13205_2019_1898_MOESM6_ESM.pptx
Supplementary Fig. S5 Agarose gel electrophoresis of total RNA isolated from the mature flower of four Dendrobium hybrid flowers. Lanes 1–3: Dendrobium Burmese ruby × Dendrobium Mae-klong River; Lanes 4–6: Dendrobium Burana Jade × [Dendrobium Bertha Chong × Dendrobium Imelda Romualdez]), Lanes 7–9: Dendrobium Trudy Brandt × Dendrobium Udom Blue Angel, Lanes 10–12: Dendrobium Aridang × Dendrobium Burana Sundae and Lane 13: 1 kb DNA Ladder (Promega, USA)
Rights and permissions
About this article
Cite this article
Khairul-Anuar, MA., Mazumdar, P., Lau, SE. et al. High-quality RNA isolation from pigment-rich Dendrobium flowers. 3 Biotech 9, 371 (2019). https://doi.org/10.1007/s13205-019-1898-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1898-y