Abstract
Tea, a beverage crop, is a rich source of polyphenols and polysaccharides which greatly attribute to its importance. However, oxidation and precipitation of these compounds during nucleic acids extraction is a limitation to molecular biology and genomic studies. On isolation of total RNA from root tissue using established protocols, difficulties were encountered in terms of purity and quantity of isolated RNA or some of the methods were time-consuming and also yields were low. The present communication combines a phenol-based RNA isolation protocol with a cetyltrimethylammonium bromide-based procedure with appropriate modifications. This protocol successfully isolated RNA from tap root tissue in 2–3 h as compared with 16 h reported by the previous method. Also, RNA yield was higher by more than fourfold. The RNA isolated by this protocol was successfully used for downstream applications such as RT-PCR and the construction of suppression subtractive hybridization library. The developed protocol worked well with other plant tissue with high polyphenols and polysaccharides contents.


Similar content being viewed by others
References
Sharma, P., & Kumar, S. (2005). Differential display-mediated identification of three drought-responsive expression sequence tags in tea (Camellia sinensis (L.) O. Kuntze). Journal of Biosciences, 30, 231–235.
Singh, K., Rani, A., Kumar, S., Sood, P., Mahajan, M., Yadav, S. K., et al. (2008). An early gene of the flavonoid pathway, flavanone 3-hydroxylase, exhibits a positive relationship with the concentration of catechins in tea (Camellia sinensis). Tree Physiology, 28, 1349–1356.
Rani, A., Singh, K., Sood, P., Kumar, S., & Ahuja, P. S. (2009). p-Coumarate:CoA ligase as a key gene in the yield of catechins in tea [Camellia sinensis (L.) O. Kuntze]. Functional and Integrative Genomics, 9, 271–275.
Vyas, D., Kumar, S., & Ahuja, P. S. (2007). Tea (Camellia sinensis) clones with shorter periods of winter dormancy exhibit lower accumulation of reactive oxygen species. Tree Physiology, 27, 1253–1259.
Paul, A., & Kumar, S. (2011). Responses to winter dormancy, temperature, and plant hormones share gene networks. Functional and Integrative Genomics, 11(4), 659–664. doi:10.1007/s10142-011-0233-4.
Paul, A., Lal, L., Ahuja, P. S., & Kumar, S. (2011). Alpha-tubulin (CsTUA) up-regulation during winter dormancy is a low temperature inducible gene in tea [Camellia sinensis (L.) O. Kuntze]. Molecular Biology Reports. doi:10.1007/s11033-011-1121-7.
Yang, C. S., & Landau, J. M. (2000). Effect of tea consumption on nutrition and health. Journal of Nutrition, 130, 2409–2412.
Wang, D. F., Xie, X. F., Cai, C. Y., & Yang, M. (1995). Analysis of pharmacological components of coarse tea curing diabetes. Chinese Traditional and Herbal Drugs, 26, 255–257.
Loomis, W. (1974). Overcoming problems of phenolic and quinones in the isolation of plant enzymes and organelles. Methods in Enzymology, 31, 528–545.
Chang, S., Puryear, J., & Cairney, J. (1993). A simple and efficient method for isolating RNA from pine. Plant Molecular Biology Reporter, 11, 113–116.
Salzman, R. A., Fujita, T., Zhu-Salzman, K., Hasegawa, P. M., & Bressan, R. A. (1999). An improved method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Molecular Biology Reporter, 17, 11–17.
Lal, L., Sahoo, R., Gupta, R. K., Sharma, P., & Kumar, S. (2001). RNA isolation from high-phenolic tea leaves and apical buds. Plant Molecular Biology Reporter, 19, 181a–181f.
Japelaghi, R. H., Haddad, R., & Garoosi, G. A. (2011). Rapid and efficient isolation of high quality nucleic acids from plant tissues rich in polyphenols and polysaccharides. Molecular Biotechnology, 49, 129–137.
Reid, K. E., Olsson, N., Schlosser, J., Peng, F., & Lund, S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biology, 6, 27.
Moser, C., Gatto, P., Moser, M., Pindo, M., & Velasco, R. (2004). Isolation of functional RNA from small amounts of different grape and apple tissues. Molecular Biotechnology, 26, 95–99.
Ghawana, S., Paul, A., Kumar, H., Kumar, A., Singh, H., Bhardwaj, P. K., et al. (2011). An RNA isolation system for plant tissues rich in secondary metabolites. BMC Research Notes, 4, 85.
Wang, X., Xiao, H., Chen, G., Zhao, X., Huang, C., Chen, C., et al. (2011). Isolation of high-quality RNA from Reaumuria soongorica, a desert plant rich in secondary metabolites. Molecular Biotechnology, 48, 165–172.
Chawla, R., Arora, R., Kumar, R., Sharma, A., Prasad, J., Singh, S., et al. (2005). Antioxidant activity of fractionated extracts of rhizomes of high-altitude Podophyllum hexandrum: Role in radiation protection. Molecular and Cellular Biochemistry, 273, 193–208.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press.
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., et al. (2006). The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology, 7, 3.
Singh, K., Raizada, J., Bhardwaj, P., Ghawana, S., Rani, A., Singh, H., et al. (2004). 26S rRNA-based internal control gene primer pair for reverse transcription polymerase chain reaction-based quantitative expression studies in diverse plant species. Analytical Biochemistry, 335, 330–333.
Gasic, K., Hernandez, A., & Korban, S. S. (2004). RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Molecular Biology Reporter, 22, 437a–437g.
Wise, M. J., & Tunnacliffe, A. (2004). POPP the question: What do LEA proteins do? Trends in Plant Science, 9, 13–17.
Acknowledgments
Authors thank the Council of Scientific and Industrial Research (CSIR), India for funding the project through Supra Institutional project entitled “high value products from agroforestry resources from the Himalayan region and improving productivity and quality of product development, including evaluation facility for nutraceutical/value added products (SIP-003)” and “pathway engineering and system biology approach toward homologous and heterologous expression of high value phytoceuticals (NWP-008)”. Authors thank the Director, CSIR-Institute of Himalayan Bioresource Technology (IHBT) for providing necessary facilities for the work. RCM is grateful to CSIR and the Academy of Sciences for Developing Countries (TWAS), Italy for the award of Postgraduate Fellowship. AP thanks CSIR for awarding Junior/Senior Research Fellowships. AK gratefully acknowledges CSIR for providing assistantship. Thanks are due to Mr. Mohit Kumar Swarnkar for help in Bioanalyzer evaluation and to Mr. Digvijay Singh Naruka for help in plasmid isolation and sequencing. Manuscript represents IHBT communication number 2249.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12033_2011_9476_MOESM1_ESM.ppt
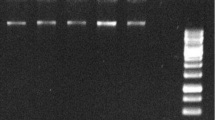
Supplementary Fig. 1 Amenability of RNA isolated from rhizome of P. hexandrum for downstream applications as evidenced using RT-PCR based amplification of 26S rRNA. Corresponding RNA is shown in Fig. 1b, lane 5; RNA was absent in lanes 1-4 in Fig. 1b and hence PCR data is not shown for those lanes (there was no amplification). (PPT 492 kb)
12033_2011_9476_MOESM2_ESM.ppt
Supplementary Fig. 2 Integrated density value (IDV) of the amplicons obtained for plants grown in the field (panel a; corresponding amplicons are shown in Fig. 2c), and green house (panel b; corresponding amplicons are shown in Fig. 2d). Plants in the green house were subjected to drought stress by withholding water as detailed in materials and methods section. IDV was measured using Alpha DigiDoc 1000 software as detailed in materials and methods section. TAB (apical bud and the associated two leaves); ML (mature leaf, pooled tissue at node positions 4th, 5th, and 6th with reference to the apical bud); FB, flower bud; C, control; DS, drought stressed. Other details are mentioned in the legend of Fig. 2. (PPT 227 kb)
Rights and permissions
About this article
Cite this article
Muoki, R.C., Paul, A., Kumari, A. et al. An Improved Protocol for the Isolation of RNA from Roots of Tea (Camellia sinensis (L.) O. Kuntze). Mol Biotechnol 52, 82–88 (2012). https://doi.org/10.1007/s12033-011-9476-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9476-5