Abstract
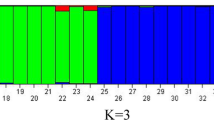
The main aim of this study is to assess the potentiality of SSR markers for the identification of the cross-species transferability frequency in a large set of the diverse genome types of wild relative rice along with cultivated rice. Here, we used 18 different rice genotypes representing nine different genome types with 70 SSR markers to investigate the potentiality of cross-species transferability rate. The overall cross-species transferability of SSR markers across the 18 rice genotypes ranged from 38.9% (RM280 and RM447) to 100% (RM490, RM318, RM279, RM18877 and RM20033, RM19303) with an average of 76.58%. Also, cross-species transferability across chromosome ranged from 54.4% (chromosome 4) to 86.5% (chromosome 2) with an average of 74.35%. The polymorphism information content of the markers varied from 0.198 (RM263) to 0.868 (RM510) with a mean of 0.549 ± 0.153, showing high discriminatory power. The highest rate of cross-transferability was observed in O. rufipogon (97%), The highest rate of cross-species transferability was in O. rufipogon (97.00%), followed by O. glaberrima (94.20%), O. nivara (92.80%), Swarna (92.80%), O. longistaminata (91.40%), O. eichingeri (90%), O. barthii (88.50%), O. alta (82.80%), O. australiensis (77.10%), O. grandiglumis (74.20%), O. officinalis (74.20%), Zizania latifolia (70.00%), O. latifolia (68.50%), O. brachyantha (62.80%), Leersia perrieri (57.10%) and O. ridleyi (41.40%) with least in O. coarctata (28.50%). A total of 341 alleles from 70 loci were detected with the number of alleles per locus ranged from 2 to 12. Based on dendrogram analysis, the AA genome groups was separated as distinct group from the rest of the genome types. Similarly, principal coordinate analysis and structure analysis clearly separated the AA genome type from the rest of the genome types. Through the analysis of molecular variance, more variance (51%) was observed among the individual, whereas less (14%) was observed among the population. Thus, our findings may offer a valuable resource for studying the genetic diversity and relationship to facilitate the understanding of the complex mechanism of the origin and evolutionary processes of different Oryza species and wild relative rice.




Similar content being viewed by others
References
Alexander LW, Thammina CS, Kramer M (2018) Cross-transferability of SSR markers in Osmanthus. Genet Resour Crop Evol 65(1):125–136
Brondani C, Rangel PHN, Borba TCO, Brondani RPV (2003) Transferability of microsatellite and sequence tagged site markers in Oryza species. Hereditas 138:187–192
Brondani RPV, Zucchi MI, Brondani C, Rangel PHN, Borba TCDO, Rangel PN et al (2005) Genetic structure of wild rice Oryza glumaepatula populations in three Brazilian biomes using microsatellite markers. Genetica 125:115–123
Chen J, Huang Q, Gao D, Wang J, Lang Y, Liu T et al (2013) Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun 4:1595
Choudhury B, Khan ML, Dayanandan S (2013) Genetic structure and diversity of indigenous rice varieties (Oryza sativa) in Eastern Himalayan region of Northeast India. Springerplus 2:228–237
Dayanandan S, Bawa K, Kesseli R (1997) Conservation of microsatellites among tropical trees (Leguminosae). Am J Bot 84:1658–1663
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Fernandez-Silva I, Eduardo I, Blanca J, Esteras C, Pico B, Nuez F et al (2008) Bin mapping of genomic and EST-derived SSRs in melon (Cucumis melo L.). Theor Appl Genet 118(1):139–150
Gao LZ, Zhang CH, Jia JZ, Dong YS (2005) Cross-species transferability of rice microsatellites in its wild relatives and the potential for conservation genetic studies. Genet Resour Crop Evol 52:931–940
Ge S, Sang T, Lu BR, Hong DY (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA 96:14400–14405
Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genomics 270(4):315–323
Hernández P, Dorado G, Laurie DA, Martin A, Snape JW (2001) Microsatellites and RFLP probes from maize are efficient sources of molecular markers for the biomass energy crop Miscanthus. Theor Appl Genet 102(4):616–622
Huang X, Zhao Y, Li C, Wang A, Zhao Q, Li W et al (2012) Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet 44(1):32–39
Ishii T, Hiraoka T, Kanzaki T, Akimoto M, Shishido R, Ishikawa R (2011) Evaluation of genetic variation among wild populations and local varieties of rice. Rice 4:170–177
Izawa T (2008) The process of rice domestication: a new model based on recent data. Rice 1(2):127–134
Jacquemin J, Bhatia D, Singh K, Wing RA (2013) The International Oryza Map Alignment Project: development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr Opin Plant Biol 16(2):147–156
Jena KK, Khush GS (1990) Introgression of genes from O. officinalis Wall. ex Watt to cultivated rice. O. sativa L. Theor Appl Genet 80:737–745
Khush G (2003) Productivity improvements in rice. Nutr Rev 61:S114–S116
Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS et al (2015) Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep 5:15655
Krishnan SG, Waters DLE, Henry RJ (2014) Australian wild rice reveals pre-domestication origin of polymorphism deserts in rice genome. PLoS ONE 9(6):e98843
Li G, Hu W, Qin R, Jin H, Tan G, Zhu L, He G (2008) Simple sequence repeat analyses of interspecific hybrids and MAALs of Oryza officinalis and Oryza sativa. Genetica 134(2):169–180
Liu L, Wang Y, He P, Li P, Lee J, Soltis DE, Fu C (2018) Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genomics 19(1):235
Mansueto L, Fuentes RR, Borja FN, Detras J, Abriol-Santos JM, Chebotarov D et al (2016) Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res 45(D1):D1075–D1081
Nachimuthu SVV, Muthurajan R, Duraialaguraja S, Sivakami R, Pandian BA, Ponniah G et al (2015) Analysis of population structure and genetic diversity in rice germplasm using SSR markers: an initiative towards association mapping of agronomic traits in Oryza. Rice 8:30
Nei M (1972) Genetic distance between populations. Am Nat 106(949):283–292
Ngangkham U, Parida SK, De S, Kumar AR, Singh AK, Singh NK, Mohapatra T (2010) Genic markers for Wild Abortive (WA) based male sterility and its fertility restoration in rice. Mol Breed 26:275–292
Ngangkham U, Samantaray S, Yadav MK, Kumar A, Chidambaranathan P, Katara JL (2018a) Effect of multiple allelic combinations of genes on regulating grain size in rice. Plos ONE 13(1):e0190684
Ngangkham U, Nath M, Dokku P, Mithra SA, Ramamurthy S, Singh NK et al (2018b) An EMS-induced new sequence variant, TEMS5032, in the coding region of SRS3 gene leads to shorter grain length in rice (Oryza sativa L.). J Appl Genet 59(4):377–389
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13(5):1143–1155
Orn C, Shishido R, Akimoto M, Ishikawa R, Htun TM, Nonomura KI et al (2015) Evaluation of genetic variation among wild rice populations in Cambodia. Breed Sci 65(5):430–437
Panaud O, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252(5):597–607
Patra BC, Ray S, Ngangkham U, Mohapatra T (2015) Rice. In: Upadhyaya HD, Singh M (eds) Genetic and genomic resources for grain cereals improvement. Elsevier Academic Press, Cambridge
Peakall RO, Smouse PE (2012) GenAlex 6.5: genetic analysis in Excel. Population genetic software for teaching and research. Bioinformatics 28:2537–2539
Perrier X, Jacquemoud-Collet JP (2006) DARwin software (http://darwin.cirad.fr/darwin)
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Ramiah K (1953) Rice breeding and genetics. ICAR, New Delhi
Ray S, Bose LK, Ray J, Ngangkham U, Katara JL, Samantaray S et al (2016) Development and validation of cross-transferable and polymorphic DNA markers for detecting alien genome introgression in Oryza sativa from Oryza brachyantha. Mol Genet Genomics 291(4):1783–1794
Shakiba E, Eizenga GC (2014) Unraveling the secrets of rice wild species. In: Rice-germplasm, genetics and improvement. InTech
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29(1):308–311
Song ZP, Xu X, Wang B, Chen JK, Lu BR (2003) Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theor Appl Genet 107(8):1492–1499
Song S, Tian D, Zhang Z, Hu S, Yu J (2019) Rice genomics: over the past two decades and into the future. Genom Proteomics Bioinf 1:1. https://doi.org/10.1016/j.gpb.2019.01.001
Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C et al (2018) Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet 50(2):285
Tan G, Jin H, Li G, He R, Zhu L, He G (2005) Production and characterization of a complete set of individual chromosome additions from Oryza officinalis to Oryza sativa using RFLP and GISH analyses. Theor Appl Genet 111(8):1585–1595
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277(5329):1063–1066
Tello-Ruiz MK, Stein J, Wei S, Preece J, Olson A, Naithani S et al (2015) Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res 44(D1):D1133–D1140
Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Curr Opin Plant Biol 6(2):139–146
Wang ML, Chen ZB, Barkley NA, Newman ML, Kim W, Raymer P et al (2006) Characterization of seashore Paspalum (Paspalum vaginatum Swartz) germplasm by transferred SSRs from wheat, maize and sorghum. Genet Resour Crop Evol 53(4):779–791
Wang C, Liu X, Peng S, Xu Q, Yuan X, Feng Y et al (2014) Development of novel microsatellite markers for the BBCC Oryza genome (Poaceae) using high-throughput sequencing technology. PloSone 9(3):e91826
Wu KS, Tanksley SD (1993) Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Gen Genet 241(1–2):225–235
Yadav MK, Aravindan S, Ngangkham U, Shubudhi HN, Bag MK, Totan Adak T et al (2017) Use of molecular markers in identification and characterization of resistance to rice blast in India. Plos ONE 12(4):e0176236
Yamaki S, Ohyanagi H, Yamasaki M, Eiguchi M, Miyabayashi T, Kubo T et al (2013) Development of INDEL markers to discriminate all genome types rapidly in the genus Oryza. Breed Sci 63(3):246–254
Yan Z, Wu F, Luo K, Zhao Y, Yan Q, Zhang Y et al (2017) Cross-species transferability of EST-SSR markers developed from the transcriptome of Melilotus and their application to population genetics research. Sci Rep 7(1):17959
Yeh FC (1997) Population genetic analysis of co-dominant and dominant marker and quantitative traits. Belgian J Bot 130:129–157
Yelome OI, Audenaert K, Landschoot S, Dansi A, Vanhove, Silue D et al (2018) Analysis of population structure and genetic diversity reveals gene flow and geographic patterns in cultivated rice (O. sativa and O. glaberrima) in West Africa. Euphytica 214:215
Yuan N, Sun Y, Comes HP, Fu CX, Qiu YX (2014) Understanding population structure and historical demography in a conservation context: population genetics of the endangered Kirengeshoma palmata (Hydrangeaceae). Am J Bot 101(3):521–529
Zhang QJ, Gao LZ (2017) Rapid and recent evolution of LTR retrotransposons drives rice genome evolution during the speciation of AA-genome Oryza species. G3: Genes Genom Genet 7(6):1875–1885
Zhang QJ, Zhu T, Xia EH, Shi C, Liu YL, Zhang Y et al (2014) Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc Natl Acad Sci USA 111(46):E4954–E4962
Zhao H, Yao W, Ouyang Y, Yang W, Wang G, Lian X et al (2014) RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res 43(D1):D1018–D1022
Zheng X, Pan C, Diao Y, You Y, Yang C, Hu Z (2013) Development of microsatellite markers by transcriptome sequencing in two species of Amorphophallus (Araceae). BMC Genomics 14(1):490
Zhou Q, Luo D, Ma L, Xie W, Wang Y, Wang Y, Liu Z (2016) Development and cross-species transferability of EST-SSR markers in Siberian Wildrye (Elymus sibiricus L.) using Illumina sequencing. Sci Rep 6:20549
Zhu T, Xu PZ, Liu JP, Peng S, Mo XC, Gao LZ (2014) Phylogenetic relationships and genome divergence among the AA-genome species of the genus Oryza as revealed by 53 nuclear genes and 16 intergenic regions. Mol Phylogenet Evol 70:348–361
Acknowledgments
The authors are extremely grateful to the Director, ICAR-National Rice Research Institute, Cuttack, India, for his support and facilitation for carrying out the research work successfully.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no potential competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ngangkham, U., Dash, S., Parida, M. et al. The potentiality of rice microsatellite markers in assessment of cross-species transferability and genetic diversity of rice and its wild relatives. 3 Biotech 9, 217 (2019). https://doi.org/10.1007/s13205-019-1757-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1757-x