Abstract
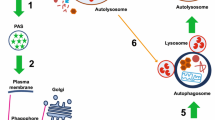
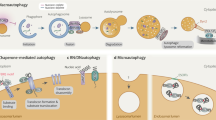
Macroautophagy (hereafter called autophagy) is a highly conserved lysosomal pathway for catabolism of intracellular material in eukaryotic cells. Autophagy is also an essential homeostatic process through which intracellular components are recycled for reuse or energy production. The extremely regulated autophagy process begins with the formation of hallmarked double membrane bound organelles called autophagosomes which in turn fuse with lysosomes called autolysosomes and finally degrade the autophagic cargos. The multistages molecular machinery of autophagy is critically orchestrated by the action of a set of the autophagy proteins (Atg) and a supreme regulator, mTOR (mechanistic target of rapamycin). However, individual stages of autophagy are mechanistically complex and partially understood. In this review, the individual stages of autophagy are dissected, and the corresponding molecular regulation is discussed in view of current scientific knowledge of autophagy. This understanding of sequential events of autophagy machinery through this review may lead to great interest in the therapeutic potential for manipulating of autophagy in established diseases.








Similar content being viewed by others
References
Abada A, Elazar Z (2014) Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep 15:839–852
Al-Bari MAA, Xu P (2020) Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann N Y Acad Sci 1467:3–20. https://doi.org/10.1111/nyas.14305
Backer JM (2016) The intricate regulation and complex functions of the class III phosphoinositide 3kinase Vps34. Biochem J 473:2251–2271
Bejarano E, Yuste A, Patel B, Stout Jr RF, Spray DC, Cuervo AM (2014) Connexins modulate autophagosome biogenesis. Nat Cell Biol 16:401–414
Biazik J, Yla-Anttila P, Vihinen H et al (2015) Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy 11:439–451
Carlsson SR, Simonsen A (2015) Membrane dynamics in autophagosome biogenesis. J Cell Sci 128:193–205
Chang J, Lee S, Blackstone C (2014) Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J Clin Invest 124(12):5249–5262
Chen K, Shi W (2016) Autophagy regulates resistance of non-small cell lung cancer cells to paclitaxel. Tumour Biol 37:10539–10544
Chen Y, Yu L (2017) Recent progress in autophagic lysosome reformation. Traffic 18(6):358–361
Chen Y, Yu L (2018) Development of research into autophagic lysosome reformation. Mol Cells 41(1):45–49
Conway O, Kirkin V (2017) Love laughs at locksmiths: Ubiquitylation of p62 unlocks its autophagy receptor potential. Cell Res 27(5):595–597
Cook KL, Soto-Pantoja DR, Abu-Asab M, Clarke PAG, Roberts DD, Clarke R (2014) Mitochondria directly donate their membrane to form autophagosomes during a novel mechanism of parkin- associated mitophagy. Cell Biosci 4:16. https://doi.org/10.1186/2045-3701-4-16
Davis S, Wang J, Zhu M (2016) Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. eLife 5. https://doi.org/10.7554/eLife.21167
Deng Z, Purtell K, Lachance V, Wold MS, Chen S, Yue Z (2017) Autophagy receptors and neurodegenerative diseases. Trends Cell Biol 27(7):491–504
Di Bartolomeo S, Corazzari M, Nazio F et al (2010) The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 191(1):155–168
Di Maio R, Barrett PJ, Hoffman EK et al (2016) Alpha-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci Transl Med 8(342):342ra378
Dikic I, Elazar Z (2018) Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19(6):349–364
Dooley HC, Razi M, Polson HE et al (2014) WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cells 55:238–252
Du W, Su QP, Chen Y et al (2016) Kinesin 1 drives autolysosome tubulation. Dev Cell 37(4):326–336
Feng YC, Backues SK, Baba M, Heo JM, Harper JW, Klionsky DJ (2016) Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy 12:648–658
Fernandez AF, Sebti S, Wei Y et al (2018) Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558(7708):136–140
Fracchiolla D, Sawa-Makarska J, Martens S (2017) Beyond Atg8 binding: the role of AIM/LIR motifs in autophagy. Autophagy 13(5):978–979
Fujioka Y, Suzuki SW, Yamamoto H, Kondo-Kakuta C, Kimura Y, Hirano H, Akada R, Inagaki F, Ohsumi Y, Noda NN (2014) Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol 21:513–521
Ge L, Wilz L, Schekman R (2015) Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment. Autophagy 11:2372–2374
Ge L, Zhang M, Kenny SJ, Liu D, Maeda M, Saito K, Mathur A, Xu K, Schekman R (2017) Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep 18:1586–1603
Ge L, Zhang M, Schekman R (2014) Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife 3:04135
Geng J, Klionsky DJ (2017) Direct quantification of autophagic flux by a single molecule-based probe. Autophagy 13:639–641
Graef M, Friedman JR, Graham C, Babu M, Nunnari J (2013) ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell 24:2918–2931
Grumati P, Dikic I (2018) Ubiquitin signaling and autophagy. J Biol Chem 293:5404–5413
Guo B, Liang Q, Li L, Hu Z, Wu F, Zhang P, Ma Y, Zhao B, Kovács AL, Zhang Z, Feng D, Chen S, Zhang H (2014) O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat Cell Biol 16(12):1215–1226
Hasegawa J, Iwamoto R, Otomo T, Nezu A, Hamasaki M, Yoshimori T (2016) Autophagosome-lysosome fusion in neurons requires INPP5E, a protein associated with Joubert syndrome. EMBO J 35(17):1853–1867
Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11:1433–1437
Hong Z, Pedersen NM, Wang L, Torgersen ML, Stenmark H, Raiborg C (2017) PtdIns3P controls mTORC1 signaling through lysosomal positioning. J Cell Biol 216(12):4217–4233
Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, Liu W (2015) Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell 57:456–466. https://doi.org/10.1016/j.molcel.2014.12.013
Hurley JH, Young LN (2017) Mechanisms of autophagy initiation. Annu Rev Biochem 86:225–244
Isakson P, Holland P, Simonsen A (2013) The role of ALFY in selective autophagy. Cell Death Differ 20(1):12–20
Itakura E, Kishi-Itakura C, Mizushima N (2012) The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151(6):1256–1269
Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ (2014) Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol 24:1314–1322
Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT (2016) Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun 7:12420
Kast DJ, Dominguez R (2017) The cytoskeleton-autophagy connection. Curr Biol 27(8):R318–R326
Kenific CM, Debnath J (2016) NBR1-dependent selective autophagy is required for efficient cell-matrix adhesion site disassembly. Autophagy 12(10):1958–1959
Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510(7503):162–166
Kraft LJ, Dowler J, Manral P, Kenworthy AK (2016) Size, organization, and dynamics of soluble SQSTM1 and LC3-SQSTM1 complexes in living cells. Autophagy 12:1660–1674
Krols M, Bultynck G, Janssens S (2016) ER-mitochondria contact sites: a new regulator of cellular calcium flux comes into play. J Cell Biol 214:367–370
Kumar S, Jain A, Farzam F (2018) Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol 217(3):997–1013
Lamark T, Svenning S, Johansen T (2017) Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem 61(6):609–624
Lamb CA, Nühlen S, Judith D, Frith D, Snijders AP, Behrends C, Tooze SA (2016) TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J 35:281–301
Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ (2015) The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524(7565):309–314
Lazarus MB, Novotny CJ, Shokat KM (2015) Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem Biol 10:257–261
Li Y, Ding W-X (2017) Impaired Rab7 and Dynamin2 block fat turnover by autophagy in alcoholic fatty livers. Hepatol Commun 1:473–476
Lin MG, Hurley JH (2016) Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol 39:61–68
Liu X, Klionsky DJ (2015) TP53INP2/DOR protein chaperones deacetylated nuclear LC3 to the cytoplasm to promote macroautophagy. Autophagy 11(9):1441–1442
Luo S, Garcia-Arencibia M, Zhao R (2012) Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell 47(3):359–370
Mackeh R, Perdiz D, Lorin S, Codogno P, Pous C (2013) Autophagy and microtubules - new story, old players. J Cell Sci 126:1071–1080
Maday S, Holzbaur ELF (2014) Autophagosome biogenesis inprimary neuronsfollows anordered and spatially regulated pathway. Dev Cell 30:71–85
Martina JA, Chen Y, Gucek M, Puertollano R (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8(6):903–914
Morishita H, Kaizuka T, Hama Y, Mizushima N (2017) A new probe to measure autophagic flux in vitro and in vivo. Autophagy 13:757–758
Munson MJ, Allen GF, Toth R et al (2015) mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J 34(17):2272–2290
Nakamura S, Yoshimori T (2017) New insights into autophagosome-lysosome fusion. J Cell Sci 130(7):1209–1216
Nakashima H, Nguyen T, Goins WF, Chiocca EA (2015) Interferon-stimulated gene 15 (ISG15) and ISG15-linked proteins can associate with members of the selective autophagic process, histone deacetylase 6 (HDAC6) and SQSTM1/p62. J Biol Chem 290(3):1485–1495
Nascimbeni AC, Giordano F, Dupont N, Grasso D, Vaccaro MI, Codogno P, Morel E (2017) ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J 36:2018–2033
Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M (2016) Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 215(6):857–874
Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pébusque MJ, Vaccaro MI, Velasco G, Dagorn JC, Iovanna JL (2009) The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell 20:870–881
Okatsu K, Kimura M, Oka T, Tanaka K, Matsuda N (2015) Unconventional PINK1 localization to the outer membrane of depolarized mitochondria drives Parkin recruitment. J Cell Sci 128(5):964–978
Olsvik HL, Lamark T, Takagi K, Larsen KB, Evjen G, Øvervatn A, Mizushima T, Johansen T (2015) FYCO1 contains a C-terminally extended, LC3A/B-preferring LC3-interacting region (LIR) motif required for efficient maturation of autophagosomes during basal autophagy. J Biol Chem 290(49):29361–29374
Olszewska DA, Lynch T (2016) PINK1, parkin, and autophagy receptors: a new model of mitophagy. Mov Disord 31(11):1628–1629
Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA (2012) Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 23:1860–1873
O'Sullivan TE, Johnson LR, Kang HH et al (2015) BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity 43(2):331–342
Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Øvervatn A, Bjørkøy G, Johansen T (2010) FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 188(2):253–269
Peng H, Yang J, Li G, You Q, Han W, Li T, Gao D, Xie X, Lee BH, du J, Hou J, Zhang T, Rao H, Huang Y, Li Q, Zeng R, Hui L, Wang H, Xia Q, Zhang X, He Y, Komatsu M, Dikic I, Finley D, Hu R (2017) Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res 27(5):657–674
Piano Mortari E, Folgiero V, Marcellini V, Romania P, Bellacchio E, D'Alicandro V, Bocci C, Carrozzo R, Martinelli D, Petrini S, Axiotis E, Farroni C, Locatelli F, Schara U, Pilz DT, Jungbluth H, Dionisi-Vici C, Carsetti R (2018) The Vici syndrome protein EPG5 regulates intracellular nucleic acid trafficking linking autophagy to innate and adaptive immunity. Autophagy 14(1):22–37
Proikas-Cezanne T, Takacs Z, Donnes P et al (2016) WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J Cell Sci 128:207–217
Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS (2016) Mechanisms and functions of lysosome positioning. J Cell Sci 129(23):4329–4339
Rabanal-Ruiz Y, Otten EG, Korolchuk VI (2017) mTORC1 as the main gateway to autophagy. Essays Biochem 61(6):565–584
Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC (2010) Plasma membrane contributes to the formationof pre-autophagosomal structures. Nat Cell Biol 12:747–757
Romero M, Sabaté-Pérez A, Francis VA, Castrillón-Rodriguez I, Díaz-Ramos Á, Sánchez-Feutrie M, Durán X, Palacín M, Moreno-Navarrete JM, Gustafson B, Hammarstedt A, Fernández-Real JM, Vendrell J, Smith U, Zorzano A (2018) TP53INP2 regulates adiposity by activating β-catenin through autophagy-dependent sequestration of GSK3β. Nat Cell Biol 20:443–454. https://doi.org/10.1038/s41556-018-0072-9
Kang R, Livesey KM, Zeh HJ et al (2010) HMGB1: a novel Beclin 1-binding protein active in autophagy. Autophagy 6(8):1209–1211. https://doi.org/10.4161/auto.6.8.13651
Sanchez-Wandelmer J, Ktistakis NT, Reggiori F (2015) ERES: sites for autophagosome biogenesis and maturation? J Cell Sci 128:185–192
Sasaki T, Lian S, Khan A, Llop JR, Samuelson AV, Chen W, Klionsky DJ, Kishi S (2017) Autolysosome biogenesis and developmental senescence are regulated by both Spns1 and v-ATPase. Autophagy 13(2):386–403
Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 169(2):361–371
Song P, Li S, Wu H, Gao R, Rao G, Wang D, Chen Z, Ma B, Wang H, Sui N, Deng H, Zhang Z, Tang T, Tan Z, Han Z, Lu T, Zhu Y, Chen Q (2016) Parkin promotes proteasomal degradation of p62: implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson’s disease. Protein Cell 7(2):114–129
Søreng K, Munson MJ, Lamb CA et al (2018) SNX18 regulates ATG9A trafficking from recycling endosomes by recruiting Dynamin-2. EMBO Rep 19(4). https://doi.org/10.15252/embr.201744837
Stadel D, Millarte V, Tillmann KD et al (2015) TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol Cell 60:89–104
Suzuki H, Kaizuka T, Mizushima N, Noda NN (2015) Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat Struct Mol Biol 22(7):572–580
Tait SW, Ichim G, Green DR (2014) Die another way--non-apoptotic mechanisms of cell death. J Cell Sci 127(Pt 10):2135–2144
Tanida I, Ueno T, Kominami E (2004) LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 36:2503–2518
Terawaki S, Camosseto V, Pierre P, Gatti E (2016) RUFY4: Immunity piggybacking on autophagy? Autophagy 12(3):598–600
Tumbarello DA, Manna PT, Allen M, Bycroft M, Arden SD, Kendrick-Jones J, Buss F (2015) The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of Salmonella Typhimurium by autophagy. PLoS Pathog 11(10):e1005174
Wang H, Sun HQ, Zhu X, Zhang L, Albanesi J, Levine B, Yin H (2015) GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc Natl Acad Sci U S A 112(22):7015–7020
Wang Z, Miao G, Xue X, Guo X, Yuan C, Wang Z, Zhang G, Chen Y, Feng D, Hu J, Zhang H (2016) The Vici syndrome protein EPG5 is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol Cell 63(5):781–795
Wei Y, Liu M, Li X, Liu J, Li H (2018) Origin of the autophagosome membrane in mammals. Biomed Res Int 2018:1012789–1012789. https://doi.org/10.1155/2018/1012789
Wen X, Klionsky DJ (2016) An overview of macroautophagy in yeast. J Mol Biol 428(9 Pt a):1681–1699
Wijdeven RH, Janssen H, Nahidiazar L, Janssen L, Jalink K, Berlin I, Neefjes J (2016) Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat Commun 7:11808
Li X, Wu X-Q, Deng R, Li DD, Tang J, Chen WD, Chen JH, Ji J, Jiao L, Jiang S, Yang F, Feng GK, Senthilkumar R, Yue F, Zhang HL, Wu RY, Yu Y, Xu XL, Mai J, Li ZL, Peng XD, Huang Y, Huang X, Ma NF, Tao Q, Zeng YX, Zhu XF (2017) CaMKII-mediated Beclin 1 phosphorylation regulates autophagy that promotes degradation of Id and neuroblastoma cell differentiation. Nat Commun 8:1159. https://doi.org/10.1038/s41467-017-01272-2
You Z, Xu Y, Wan W, Zhou L, Li J, Zhou T, Shi Y, Liu W (2019) TP53INP2 contributes to autophagosome formation by promoting LC3-ATG7 interaction. Autophagy 15(8):1309–1321. https://doi.org/10.1080/15548627.2019.1580510
Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU (2006) Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 8:1124–1132
Yu Y, Zhao J (2019) Modulated autophagy by microRNAs in osteoarthritis chondrocytes. Biomed Res Int 2019:1484152
Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A (2009) DAP-kinase-mediated phosphorylation on the BH3 domain of Beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 10:285–292
Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 11:468–476
Zientara-Rytter K, Subramani S (2018) AIM/LIR-based fluorescent sensors-new tools to monitor mAtg8 functions. Autophagy 14(6):1074–1078. https://doi.org/10.1080/15548627.2018.1454238
Acknowledgements
The author gives special thanks to the Institute of Biophysics, Chinese Academy of Sciences (CAS), Beijing, China, for technical support of this manuscript. The author strongly apologizes to the investigators for not citing here all original publications due to space limitations.
Author information
Authors and Affiliations
Contributions
AAB contributed in the writing, editing, and revised of whole manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no competing interests.
Ethics approval and consent to participate
The MS is a review paper only and does not involve human participants, human data, or human tissue. So, ethics approval and consent to participate are “Not applicable”.
Consent for publication
“Not applicable”.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key points
• Autophagy is a conserved lysosomal pathway in eukaryotic cells.
• Multistages of autophagy are orchestrated by a set of autophagy proteins.
• Regulation of individual stages of autophagy is complex to understand.
• This review may lead to the treatment options for established diseases.
Rights and permissions
About this article
Cite this article
Al-Bari, M.A.A. A current view of molecular dissection in autophagy machinery. J Physiol Biochem 76, 357–372 (2020). https://doi.org/10.1007/s13105-020-00746-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13105-020-00746-0