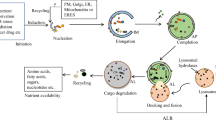
Abstract
‘Autophagy’ refers to an evolutionarily conserved process through which cellular contents, such as damaged organelles and protein aggregates, are delivered to lysosomes for degradation. Different forms of autophagy have been described on the basis of the nature of the cargoes and the means used to deliver them to lysosomes. At present, the prevailing categories of autophagy in mammalian cells are macroautophagy, microautophagy and chaperone-mediated autophagy. The molecular mechanisms and biological functions of macroautophagy and chaperone-mediated autophagy have been extensively studied, but microautophagy has received much less attention. In recent years, there has been a growth in research on microautophagy, first in yeast and then in mammalian cells. Here we review this form of autophagy, focusing on selective forms of microautophagy. We also discuss the upstream regulatory mechanisms, the crosstalk between macroautophagy and microautophagy, and the functional implications of microautophagy in diseases such as cancer and neurodegenerative disorders in humans. Future research into microautophagy will provide opportunities to develop novel interventional strategies for autophagy- and lysosome-related diseases.




Similar content being viewed by others
References
Klionsky, D. J. Autophagy revisited: a conversation with Christian de Duve. Autophagy 4, 740–743 (2008).
Marzella, L., Ahlberg, J. & Glaumann, H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J. Cell Biol. 93, 144–154 (1982).
Kovács, A. L., Reith, A. & Seglen, P. O. Accumulation of autophagosomes after inhibition of hepatocytic protein degradation by vinblastine, leupeptin or a lysosomotropic amine. Exp. Cell Res. 137, 191–201 (1982).
Klionsky, D. J. et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545 (2003).
Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 24, 9–23 (2014).
Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20, 521–527 (2018).
Feng, Y., He, D., Yao, Z. & Klionsky, D. J. The machinery of macroautophagy. Cell Res. 24, 24–41 (2014).
Kaushik, S. & Cuervo, A. M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018).
Bourdenx, M., Gavathiotis, E. & Cuervo, A. M. Chaperone-mediated autophagy: a gatekeeper of neuronal proteostasis. Autophagy 17, 2040–2042 (2021).
Mijaljica, D., Prescott, M. & Devenish, R. J. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy 7, 673–682 (2011).
Ahlberg, J., Marzella, L. & Glaumann, H. Uptake and degradation of proteins by isolated rat liver lysosomes. Suggestion of a microautophagic pathway of proteolysis. Lab. Invest. 47, 523–532 (1982).
Marzella, L., Ahlberg, J. & Glaumann, H. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 36, 219–234 (1981).
Ahlberg, J. & Glaumann, H. Uptake–microautophagy–and degradation of exogenous proteins by isolated rat liver lysosomes. Effects of pH, ATP, and inhibitors of proteolysis. Exp. Mol. Pathol. 42, 78–88 (1985).
Ahlberg, J., Berkenstam, A., Henell, F. & Glaumann, H. Degradation of short and long lived proteins in isolated rat liver lysosomes. Effects of pH, temperature, and proteolytic inhibitors. J. Biol. Chem. 260, 5847–5854 (1985).
de Waal, E. J., Vreeling-Sindelarova, H., Schellens, J. P., Houtkooper, J. M. & James, J. Quantitative changes in the lysosomal vacuolar system of rat hepatocytes during short-term starvation. A morphometric analysis with special reference to macro- and microautophagy. Cell Tissue Res. 243, 641–648 (1986).
Mortimore, G. E., Lardeux, B. R. & Adams, C. E. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J. Biol. Chem. 263, 2506–2512 (1988).
Mortimore, G. E., Hutson, N. J. & Surmacz, C. A. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc. Natl Acad. Sci. USA 80, 2179–2183 (1983).
Oku, M. & Sakai, Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays 40, e1800008 (2018).
Uttenweiler, A. & Mayer, A. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol. Biol. 445, 245–259 (2008).
Schuck, S., Gallagher, C. M. & Walter, P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127, 4078–4088 (2014).
Shen, H. M. & Mizushima, N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem. Sci. 39, 61–71 (2014).
Zhao, Y. G., Codogno, P. & Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-021-00392-4 (2021).
Kirkin, V. & Rogov, V. V. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell 76, 268–285 (2019).
Anding, A. L. & Baehrecke, E. H. Cleaning house: selective autophagy of organelles. Dev. Cell 41, 10–22 (2017).
Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017).
Quiles, J. M. & Gustafsson, A. B. Mitochondrial quality control and cellular proteostasis: two sides of the same coin. Front. Physiol. 11, 515 (2020).
Ni, H. M., Williams, J. A. & Ding, W. X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 4, 6–13 (2015).
Pickles, S., Vigie, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 (2018).
Wang, L., Qi, H., Tang, Y. & Shen, H.-M. Post-translational modifications of key machinery in the control of mitophagy. Trends Biochem. Sci. 45, 58–75 (2020).
Kissová, I. et al. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 3, 329–336 (2007).
Kissová, I., Deffieu, M., Manon, S. & Camougrand, N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 279, 39068–39074 (2004).
Welter, E. et al. Uth1 is a mitochondrial inner membrane protein dispensable for post-log-phase and rapamycin-induced mitophagy. FEBS J. 280, 4970–4982 (2013).
Lemasters, J. J. Variants of mitochondrial autophagy: types 1 and 2 mitophagy and micromitophagy (type 3). Redox Biol. 2, 749–754 (2014).
Lemasters, J. J. & Zhong, Z. Mitophagy in hepatocytes: types, initiators and role in adaptive ethanol metabolism. Liver Res. 2, 125–132 (2018).
Miyamoto, Y. et al. Possible existence of lysosome-like organella within mitochondria and its role in mitochondrial quality control. PLoS ONE 6, e16054 (2011).
Kitamura, N. et al. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS ONE 6, e16060 (2011).
Nakamura, Y. et al. BNIP3 and NIX mediate Mieap-induced accumulation of lysosomal proteins within mitochondria. PLoS ONE 7, e30767 (2012). This study reveals that SPATA18 interacts with BNIP3 or BNIP3L to induce MALM in a reactive oxygen species-dependent manner, leading to the elimination of oxidized mitochondrial proteins.
Nakamura, Y. & Arakawa, H. Discovery of Mieap-regulated mitochondrial quality control as a new function of tumor suppressor p53. Cancer Sci. 108, 809–817 (2017).
Wong, Y. C., Ysselstein, D. & Krainc, D. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 (2018).
Bhattacharya, A. & Qi, L. ER-associated degradation in health and disease - from substrate to organism. J. Cell Sci. 132, jcs232850 (2019).
Chino, H. & Mizushima, N. ER-Phagy: quality control and turnover of endoplasmic reticulum. Trends Cell Biol. 30, 384–398 (2020).
Schafer, J. A. et al. ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J. 39, e102586 (2020).
Xu, F. et al. COPII mitigates ER stress by promoting formation of ER whorls. Cell Res. 31, 141–156 (2021).
Liao, Y., Duan, B., Zhang, Y., Zhang, X. & Xia, B. Excessive ER-phagy mediated by the autophagy receptor FAM134B results in ER stress, the unfolded protein response, and cell death in HeLa cells. J. Biol. Chem. 294, 20009–20023 (2019).
Omari, S. et al. Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc. Natl Acad. Sci. USA 115, E10099–E10108 (2018). This study shows that microautophagy initiated at ERESs modified with ubiquitin, LC3 and p62 degrades a subpopulation of procollagen.
De Leonibus, C., Cinque, L. & Settembre, C. Emerging lysosomal pathways for quality control at the endoplasmic reticulum. FEBS Lett. 593, 2319–2329 (2019).
Gorrell, L., Omari, S., Makareeva, E. & Leikin, S. Noncanonical ER-Golgi trafficking and autophagy of endogenous procollagen in osteoblasts. Cell Mol. Life Sci. 78, 8283–8300 (2021).
Fregno, I. et al. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 37 https://doi.org/10.15252/embj.201899259 (2018). This study demonstrates that ERLAD relies on ER-derived, single-membrane vesicles to deliver misfolded proteins to endolysosomes for degradation, which is dependent on RETREG1, LC3 lipidation machinery and the SNARE complex.
Fregno, I. & Molinari, M. Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit. Rev. Biochem. Mol. Biol. 54, 153–163 (2019).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Loi, M., Raimondi, A., Morone, D. & Molinari, M. ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat. Commun. 10, 5058 (2019). This study shows that during recovery from ER stress, SEC62 recruits lipidated LC3 via its LIR onto ER subdomains and then the ER-derived vesicles containing fragmented ER are delivered to endolysosomes for degradation in a VPS4- and ESCRT-III-dependent manner.
Papandreou, M. E. & Tavernarakis, N. Nucleophagy: from homeostasis to disease. Cell Death Differ. 26, 630–639 (2019).
Otto, F. B. & Thumm, M. Mechanistic dissection of macro- and micronucleophagy. Autophagy 17, 626–639 (2021).
Roberts, P. et al. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 (2003).
Kvam, E. & Goldfarb, D. S. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy 3, 85–92 (2007).
Mijaljica, D., Prescott, M. & Devenish, R. J. The intricacy of nuclear membrane dynamics during nucleophagy. Nucleus 1, 213–223 (2010).
Mostofa, M. G. et al. rDNA Condensation promotes rDNA separation from nucleolar proteins degraded for nucleophagy after TORC1 inactivation. Cell Rep. 28, 3423–3434.e3422 (2019).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA 114, E4612–E4620 (2017).
Gratia, M. et al. Bloom syndrome protein restrains innate immune sensing of micronuclei by cGAS. J. Exp. Med. 216, 1199–1213 (2019).
Zhao, M. et al. CGAS is a micronucleophagy receptor for the clearance of micronuclei. Autophagy https://doi.org/10.1080/15548627.2021.1899440 (2021).
Fujiwara, Y. et al. Discovery of a novel type of autophagy targeting RNA. Autophagy 9, 403–409 (2013).
Fujiwara, Y. et al. Direct uptake and degradation of DNA by lysosomes. Autophagy 9, 1167–1171 (2013).
Fujiwara, Y., Hase, K., Wada, K. & Kabuta, T. An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding. Biochem. Biophys. Res. Commun. 460, 281–286 (2015).
Aizawa, S. et al. Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy 12, 565–578 (2016).
Aizawa, S. et al. Lysosomal membrane protein SIDT2 mediates the direct uptake of DNA by lysosomes. Autophagy 13, 218–222 (2017).
Hase, K. et al. Cytosolic domain of SIDT2 carries an arginine-rich motif that binds to RNA/DNA and is important for the direct transport of nucleic acids into lysosomes. Autophagy https://doi.org/10.1080/15548627.2020.1712109 (2020). This study reveals that SIDT2 directly binds RNA and DNA via an arginine-rich motif to mediate RN/DNautophagy. SIDT2 can also interact with exon 1 of the HTT transcript, promote the degradation of HTT mRNA and reduce the levels of HTT aggregates.
Hasegawa, J., Maejima, I., Iwamoto, R. & Yoshimori, T. Selective autophagy: lysophagy. Methods 75, 128–132 (2015).
Papadopoulos, C., Kravic, B. & Meyer, H. Repair or lysophagy: dealing with damaged lysosomes. J. Mol. Biol. 432, 231–239 (2020).
Lee, C., Lamech, L., Johns, E. & Overholtzer, M. Selective lysosome membrane turnover is induced by nutrient starvation. Dev. Cell 55, 289–297.e284 (2020).
Heckmann, B. L. & Green, D. R. LC3-associated phagocytosis at a glance. J. Cell Sci. https://doi.org/10.1242/jcs.222984 (2019).
Zhang, W. et al. A conserved ubiquitin- and ESCRT-dependent pathway internalizes human lysosomal membrane proteins for degradation. PLoS Biol. 19, e3001361 (2021).
He, C. W. et al. Membrane recruitment of Atg8 by Hfl1 facilitates turnover of vacuolar membrane proteins in yeast cells approaching stationary phase. BMC Biol. 19, 117 (2021).
Zechner, R., Madeo, F. & Kratky, D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18, 671–684 (2017).
Singh, R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009).
Kounakis, K., Chaniotakis, M., Markaki, M. & Tavernarakis, N. Emerging roles of lipophagy in health and disease. Front. Cell Dev. Biol. 7, 185 (2019).
Schulze, R. J., Sathyanarayan, A. & Mashek, D. G. Breaking fat: the regulation and mechanisms of lipophagy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1178–1187 (2017).
Liu, K. & Czaja, M. J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 20, 3–11 (2013).
Graef, M. Lipid droplet-mediated lipid and protein homeostasis in budding yeast. FEBS Lett. 592, 1291–1303 (2018).
Wang, C. W. Lipid droplet dynamics in budding yeast. Cell Mol. Life Sci. 72, 2677–2695 (2015).
van Zutphen, T. et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 25, 290–301 (2014).
Wang, C. W., Miao, Y. H. & Chang, Y. S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206, 357–366 (2014).
Vevea, J. D. et al. Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599 (2015).
Seo, A. Y. et al. AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. eLife https://doi.org/10.7554/eLife.21690 (2017).
Kurokawa, Y. et al. Microautophagy in the yeast vacuole depends on the activities of phosphatidylinositol 4-kinases, Stt4p and Pik1p. Biochim. Biophys. Acta Biomembr. https://doi.org/10.1016/j.bbamem.2020.183416 (2020).
Schulze, R. J. et al. Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc. Natl Acad. Sci. USA 117, 32443–32452 (2020). This study shows that under nutrient-limited conditions, the size of large LDs is reduced by PNPLA2 and smaller LDs directly interact with lysosomes for degradation in the absence of ATG proteins and LAMP2A.
Germain, K. & Kim, P. K. Pexophagy: a model for selective autophagy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21020578 (2020).
Cho, D. H., Kim, Y. S., Jo, D. S., Choe, S. K. & Jo, E. K. Pexophagy: molecular mechanisms and implications for health and diseases. Mol. Cell 41, 55–64 (2018).
Mukaiyama, H. et al. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cell 7, 75–90 (2002).
Sakai, Y., Koller, A., Rangell, L. K., Keller, G. A. & Subramani, S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J. Cell Biol. 141, 625–636 (1998).
Titorenko, V. I., Keizer, I., Harder, W. & Veenhuis, M. Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J. Bacteriol. 177, 357–363 (1995).
Yuan, W., Stromhaug, P. E. & Dunn, W. A. Jr. Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol. Biol. Cell 10, 1353–1366 (1999).
Manjithaya, R., Nazarko, T. Y., Farré, J. C. & Subramani, S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 584, 1367–1373 (2010).
Sakai, Y., Oku, M., van der Klei, I. J. & Kiel, J. A. Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta 1763, 1767–1775 (2006).
Farré, J. C. & Subramani, S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 14, 515–523 (2004).
Mukaiyama, H. et al. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58–70 (2004).
Chang, T. et al. PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol. Biol. Cell 16, 4941–4953 (2005).
Farré, J. C., Manjithaya, R., Mathewson, R. D. & Subramani, S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376 (2008).
Tuttle, D. L. & Dunn, W. A. Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 108, 25–35 (1995).
Sahu, R. et al. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20, 131–139 (2011). This study shows that eMI can be mediated by HSPA8 to degrade cytosolic proteins.
Chauhan, A. S. et al. Trafficking of a multifunctional protein by endosomal microautophagy: linking two independent unconventional secretory pathways. FASEB J. 33, 5626–5640 (2019).
Mesquita, A., Glenn, J. & Jenny, A. Differential activation of eMI by distinct forms of cellular stress. Autophagy https://doi.org/10.1080/15548627.2020.1783833 (2020).
Mukherjee, A., Patel, B., Koga, H., Cuervo, A. M. & Jenny, A. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 12, 1984–1999 (2016).
Huotari, J. & Helenius, A. Endosome maturation. EMBO J. 30, 3481–3500 (2011).
Lefebvre, C., Legouis, R. & Culetto, E. ESCRT and autophagies: endosomal functions and beyond. Semin. Cell Dev. Biol. 74, 21–28 (2018).
Xu, J., Camfield, R. & Gorski, S. M. The interplay between exosomes and autophagy - partners in crime. J. Cell Sci. https://doi.org/10.1242/jcs.215210 (2018).
Müller, M. et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. eLife 4, e07736 (2015).
Morozova, K. et al. Structural and biological interaction of hsc-70 protein with phosphatidylserine in endosomal microautophagy. J. Biol. Chem. 291, 18096–18106 (2016).
Uytterhoeven, V. et al. Hsc70-4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron 88, 735–748 (2015). This study demonstrates that in Drosophila spp. HSPA8 might deform the endosome membrane to promote eMI and neurotransmitter release.
Kaushik, S. & Cuervo, A. M. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 22, 407–417 (2012).
Liu, X. M. et al. ESCRTs cooperate with a selective autophagy receptor to mediate vacuolar targeting of soluble cargos. Mol. Cell 59, 1035–1042 (2015).
Arakhamia, T. et al. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 180, 633–644.e612 (2020).
Wang, Y. et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 18, 4153–4170 (2009).
Caballero, B. et al. Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat. Commun. 12, 2238 (2021). This study reveals that acetylation of MAPT reduces its degradation by CMA, while promoting its engagement by eMI.
Eskelinen, E. L. et al. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic 6, 1058–1061 (2005).
Lescat, L. et al. Chaperone-mediated autophagy in the light of evolution: insight from fish. Mol. Biol. Evol. 37, 2887–2899 (2020).
Oku, M. et al. Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 216, 3263–3274 (2017).
Yang, X. et al. TORC1 regulates vacuole membrane composition through ubiquitin- and ESCRT-dependent microautophagy. J. Cell Biol. https://doi.org/10.1083/jcb.201902127 (2020).
Mejlvang, J. et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol. 217, 3640–3655 (2018). This study shows that upon amino acid starvation, multiple autophagy receptors, including SQSTM1, NBR1, TAX1BP1 and CALCOCO2, are degraded through endolosomes, and this process is dependent on ESCRT-III and VPS4 rather than macroautophagy, CMA and HSPA8.
Linares, J. F. et al. PKCλ/ι inhibition activates an ULK2-mediated interferon response to repress tumorigenesis. Mol. Cell https://doi.org/10.1016/j.molcel.2021.08.039 (2021).
Zellner, S., Schifferer, M. & Behrends, C. Systematically defining selective autophagy receptor-specific cargo using autophagosome content profiling. Mol. Cell 81, 1337–1354.e1338 (2021).
Deguine, J. & Barton, G. M. MyD88: a central player in innate immune signaling. F1000Prime Rep. 6, 97 (2014).
Into, T. et al. Basal autophagy prevents autoactivation or enhancement of inflammatory signals by targeting monomeric MyD88. Sci. Rep. 7, 1009 (2017).
Galluzzi, L., Spranger, S., Fuchs, E. & López-Soto, A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 29, 44–65 (2019).
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Albrecht, L. V., Ploper, D., Tejeda-Muñoz, N. & De Robertis, E. M. Arginine methylation is required for canonical Wnt signaling and endolysosomal trafficking. Proc. Natl Acad. Sci. USA 115, E5317–E5325 (2018). This study demonstrates that for WNT signalling induction, cytosolic proteins are methylated by PRMT1, then phophorylated by GSK3 and ubiquitylated by E3 ligases. The modified proteins with GSK3 and PRMT1 are sequestrated and degraded through endolysosomes in an ESCRT machinery-dependent manner.
Taelman, V. F. et al. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143, 1136–1148 (2010).
Albrecht, L. V., Bui, M. H. & De Robertis, E. M. Canonical Wnt is inhibited by targeting one-carbon metabolism through methotrexate or methionine deprivation. Proc. Natl Acad. Sci. USA 116, 2987–2995 (2019).
Moon, Y. A. et al. The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab. 15, 240–246 (2012).
Zheng, Z. G. et al. Discovery of a potent SCAP degrader that ameliorates HFD-induced obesity, hyperlipidemia and insulin resistance via an autophagy-independent lysosomal pathway. Autophagy 17, 1592–1613 (2021).
Ciechanover, A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79–87 (2005).
Kwon, Y. T. & Ciechanover, A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42, 873–886 (2017).
Collins, G. A. & Goldberg, A. L. The logic of the 26S proteasome. Cell 169, 792–806 (2017).
Livneh, I., Cohen-Kaplan, V., Cohen-Rosenzweig, C., Avni, N. & Ciechanover, A. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res. 26, 869–885 (2016).
Marshall, R. S. & Vierstra, R. D. Dynamic regulation of the 26S proteasome: from synthesis to degradation. Front. Mol. Biosci. 6, 40 (2019).
Marshall, R. S. & Vierstra, R. D. Eat or be eaten: the autophagic plight of inactive 26S proteasomes. Autophagy 11, 1927–1928 (2015).
Cohen-Kaplan, V. et al. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl Acad. Sci. USA 113, E7490–E7499 (2016).
Li, J., Breker, M., Graham, M., Schuldiner, M. & Hochstrasser, M. AMPK regulates ESCRT-dependent microautophagy of proteasomes concomitant with proteasome storage granule assembly during glucose starvation. PLoS Genet. 15, e1008387 (2019).
Li, J. & Hochstrasser, M. Selective microautophagy of proteasomes is initiated by ESCRT-0 and is promoted by proteasome ubiquitylation. J. Cell Sci. https://doi.org/10.1242/jcs.259393 (2022).
Sharmin, T., Morshed, S. & Ushimaru, T. PP2A promotes ESCRT-0 complex formation on vacuolar membranes and microautophagy induction after TORC1 inactivation. Biochem. Biophys. Res. Commun. 524, 614–620 (2020).
Morshed, S., Sharmin, T. & Ushimaru, T. TORC1 regulates ESCRT-0 complex formation on the vacuolar membrane and microautophagy induction in yeast. Biochem. Biophys. Res. Commun. 522, 88–94 (2020).
Morshed, S., Tasnin, M. N. & Ushimaru, T. ESCRT machinery plays a role in microautophagy in yeast. BMC Mol. Cell Biol. 21, 70 (2020).
Hatakeyama, R. & De Virgilio, C. TORC1 specifically inhibits microautophagy through ESCRT-0. Curr. Genet. 65, 1243–1249 (2019).
Hatakeyama, R. et al. Spatially distinct pools of TORC1 balance protein homeostasis. Mol. Cell 73, 325–338.e328 (2019).
Dubouloz, F., Deloche, O., Wanke, V., Cameroni, E. & De Virgilio, C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19, 15–26 (2005).
Shaid, S., Brandts, C. H., Serve, H. & Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 20, 21–30 (2013).
Le Guerroué, F. & Youle, R. J. Ubiquitin signaling in neurodegenerative diseases: an autophagy and proteasome perspective. Cell Death Differ. 28, 439–454 (2021).
McLelland, G. L., Soubannier, V., Chen, C. X., McBride, H. M. & Fon, E. A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 33, 282–295 (2014).
Bache, K. G., Raiborg, C., Mehlum, A. & Stenmark, H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278, 12513–12521 (2003).
Katzmann, D. J., Babst, M. & Emr, S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 (2001).
Kumar, S., Jia, J. & Deretic, V. Atg8ylation as a general membrane stress and remodeling response. Cell Stress. 5, 128–142 (2021).
Martinez, J. et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl Acad. Sci. USA 108, 17396–17401 (2011).
Heckmann, B. L. et al. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell 178, 536–551.e514 (2019).
Leidal, A. M. et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199 (2020).
Florey, O., Gammoh, N., Kim, S. E., Jiang, X. & Overholtzer, M. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy 11, 88–99 (2015). The authors reveal that lipidated LC3 can be targeted to endolysosomal membranes by V-ATPase.
Fischer, T. D., Wang, C., Padman, B. S., Lazarou, M. & Youle, R. J. STING induces LC3B lipidation onto single-membrane vesicles via the V-ATPase and ATG16L1-WD40 domain. J. Cell Biol. https://doi.org/10.1083/jcb.202009128 (2020).
Nguyen, T. N. et al. ATG4 family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system. Mol. Cell 81, 2013–2030.e2019 (2021).
Agrotis, A. et al. Human ATG4 autophagy proteases counteract attachment of ubiquitin-like LC3/GABARAP proteins to other cellular proteins. J. Biol. Chem. 294, 12610–12621 (2019). This study shows that ATG3 can be modified by LC3, and the authors name this process ‘LC3ylation’.
Yim, W. W.-Y. & Mizushima, N. Lysosome biology in autophagy. Cell Discov. 6, 6 (2020).
McLelland, G. L., Lee, S. A., McBride, H. M. & Fon, E. A. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 214, 275–291 (2016). The authors demonstrate that STX17 forms a ternary SNARE complex with SNAP29 and VAMP7 to mediate the fusion of PINK1–parkin-dependent MDVs with lysosomes in a HOPS complex-dependent manner.
Huber, L. A. & Teis, D. Lysosomal signaling in control of degradation pathways. Curr. Opin. Cell Biol. 39, 8–14 (2016).
Scorrano, L. et al. Coming together to define membrane contact sites. Nat. Commun. 10, 1287 (2019).
Prinz, W. A., Toulmay, A. & Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21, 7–24 (2020).
Wong, Y. C., Kim, S., Peng, W. & Krainc, D. Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 29, 500–513 (2019).
Hao, F. et al. Rheb localized on the Golgi membrane activates lysosome-localized mTORC1 at the Golgi-lysosome contact site. J. Cell Sci. https://doi.org/10.1242/jcs.208017 (2018).
Henne, W. M. Discovery and roles of ER-endolysosomal contact sites in disease. Adv. Exp. Med. Biol. 997, 135–147 (2017).
Jin, Y., Strunk, B. S. & Weisman, L. S. Close encounters of the lysosome-peroxisome kind. Cell 161, 197–198 (2015).
Hu, Y. B., Dammer, E. B., Ren, R. J. & Wang, G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 4, 18 (2015).
Puertollano, R., Ferguson, S. M., Brugarolas, J. & Ballabio, A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. https://doi.org/10.15252/embj.201798804 (2018).
Sato, M. et al. Rapamycin activates mammalian microautophagy. J. Pharmacol. Sci. 140, 201–204 (2019).
De Falco, F., Restucci, B., Urraro, C. & Roperto, S. Microautophagy upregulation in cutaneous lymph nodes of dogs naturally infected by Leishmania infantum. Parasitol. Res. 119, 2245–2255 (2020).
Kumar, S. et al. Mammalian Atg8 proteins and the autophagy factor IRGM control mTOR and TFEB at a regulatory node critical for responses to pathogens. Nat. Cell Biol. 22, 973–985 (2020).
Nakamura, S. et al. LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury. Nat. Cell Biol. 22, 1252–1263 (2020).
Piper, R. C. & Katzmann, D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23, 519–547 (2007).
Hanson, P. I. & Cashikar, A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28, 337–362 (2012).
Frankel, E. B. & Audhya, A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 74, 4–10 (2018).
Latifkar, A., Hur, Y. H., Sanchez, J. C., Cerione, R. A. & Antonyak, M. A. New insights into extracellular vesicle biogenesis and function. J. Cell Sci. https://doi.org/10.1242/jcs.222406 (2019).
Kawamura, N. et al. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 3, 1071 (2012).
Cuttler, K., Hassan, M., Carr, J., Cloete, R. & Bardien, S. Emerging evidence implicating a role for neurexins in neurodegenerative and neuropsychiatric disorders. Open. Biol. 11, 210091 (2021).
Cuervo, A. M., Stefanis, L., Fredenburg, R., Lansbury, P. T. & Sulzer, D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 (2004).
Mak, S. K., McCormack, A. L., Manning-Bog, A. B., Cuervo, A. M. & Di Monte, D. A. Lysosomal degradation of alpha-synuclein in vivo. J. Biol. Chem. 285, 13621–13629 (2010).
Caballero, B. et al. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell https://doi.org/10.1111/acel.12692 (2018).
Park, J. S., Kim, D. H. & Yoon, S. Y. Regulation of amyloid precursor protein processing by its KFERQ motif. BMB Rep. 49, 337–342 (2016).
Huang, C. C. et al. Metabolism and mis-metabolism of the neuropathological signature protein TDP-43. J. Cell Sci. 127, 3024–3038 (2014).
Bourdenx, M. et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 184, 2696–2714.e2625 (2021).
Boassa, D. et al. Mapping the subcellular distribution of α-synuclein in neurons using genetically encoded probes for correlated light and electron microscopy: implications for Parkinson’s disease pathogenesis. J. Neurosci. 33, 2605–2615 (2013).
Sugeno, N. et al. Lys-63-linked ubiquitination by E3 ubiquitin ligase Nedd4-1 facilitates endosomal sequestration of internalized α-synuclein. J. Biol. Chem. 289, 18137–18151 (2014).
Tofaris, G. K. et al. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc. Natl Acad. Sci. USA 108, 17004–17009 (2011).
Coyne, A. N. et al. Post-transcriptional inhibition of Hsc70-4/HSPA8 expression leads to synaptic vesicle cycling defects in multiple models of ALS. Cell Rep. 21, 110–125 (2017).
Parkinson, N. et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 67, 1074–1077 (2006).
Skibinski, G. et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37, 806–808 (2005).
van der Zee, J. et al. CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum. Mol. Genet. 17, 313–322 (2008).
Mochida, G. H. et al. CHMP1A encodes an essential regulator of BMI1-INK4A in cerebellar development. Nat. Genet. 44, 1260–1264 (2012).
Rodger, C. et al. De novo VPS4A mutations cause multisystem disease with abnormal neurodevelopment. Am. J. Hum. Genet. 107, 1129–1148 (2020).
Willén, K. et al. Aβ accumulation causes MVB enlargement and is modelled by dominant negative VPS4A. Mol. Neurodegener. 12, 61 (2017).
Guo, X. et al. Amyloid β-induced redistribution of transcriptional factor EB and lysosomal dysfunction in primary microglial cells. Front. Aging Neurosci. 9, 228 (2017).
Coffey, E. E., Beckel, J. M., Laties, A. M. & Mitchell, C. H. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 263, 111–124 (2014).
Lee, J. H. et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158 (2010).
Hoffmann, A. C. et al. Extracellular aggregated alpha synuclein primarily triggers lysosomal dysfunction in neural cells prevented by trehalose. Sci. Rep. 9, 544 (2019).
Mazzulli, J. R. et al. Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146, 37–52 (2011).
Decressac, M. et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl Acad. Sci. USA 110, E1817–E1826 (2013).
Hockey, L. N. et al. Dysregulation of lysosomal morphology by pathogenic LRRK2 is corrected by TPC2 inhibition. J. Cell Sci. 128, 232–238 (2015).
Henry, A. G. et al. Pathogenic LRRK2 mutations, through increased kinase activity, produce enlarged lysosomes with reduced degradative capacity and increase ATP13A2 expression. Hum. Mol. Genet. 24, 6013–6028 (2015).
van Veen, S. et al. ATP13A2 deficiency disrupts lysosomal polyamine export. Nature 578, 419–424 (2020).
Ramirez, A. et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 38, 1184–1191 (2006).
Di Fonzo, A. et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology 68, 1557–1562 (2007).
Spataro, R. et al. Mutations in ATP13A2 (PARK9) are associated with an amyotrophic lateral sclerosis-like phenotype, implicating this locus in further phenotypic expansion. Hum. Genomics 13, 19 (2019).
Tsuji, T. et al. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. eLife https://doi.org/10.7554/eLife.25960 (2017).
Schultz, M. L., Krus, K. L. & Lieberman, A. P. Lysosome and endoplasmic reticulum quality control pathways in Niemann-Pick type C disease. Brain Res. 1649, 181–188 (2016).
Davis, O. B. et al. NPC1-mTORC1 signaling couples cholesterol sensing to organelle homeostasis and is a targetable pathway in Niemann-Pick type C. Dev. Cell 56, 260–276.e267 (2021).
Lim, C. Y. et al. ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat. Cell Biol. 21, 1206–1218 (2019).
Mulcahy Levy, J. M. & Thorburn, A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 27, 843–857 (2020).
Klionsky, D. J. et al. Autophagy in major human diseases. EMBO J. 40, e108863 (2021).
White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 12, 401–410 (2012).
Brady, C. A. et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 (2011).
Nguyen, T. A. et al. SIDT2 RNA transporter promotes lung and gastrointestinal tumor development. iScience 20, 14–24 (2019).
Yang, Y. & Bedford, M. T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 (2013).
Tejeda-Munoz, N., Albrecht, L. V., Bui, M. H. & De Robertis, E. M. Wnt canonical pathway activates macropinocytosis and lysosomal degradation of extracellular proteins. Proc. Natl Acad. Sci. USA 116, 10402–10411 (2019).
Vyas, S., Zaganjor, E. & Haigis, M. C. Mitochondria and cancer. Cell 166, 555–566 (2016).
Wallace, D. C. Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 (2012).
Tsuneki, M., Nakamura, Y., Kinjo, T., Nakanishi, R. & Arakawa, H. Mieap suppresses murine intestinal tumor via its mitochondrial quality control. Sci. Rep. 5, 12472 (2015).
Kamino, H. et al. Mieap-regulated mitochondrial quality control is frequently inactivated in human colorectal cancer. Oncogenesis 4, e181 (2016).
Okuyama, K. et al. Mieap-induced accumulation of lysosomes within mitochondria (MALM) regulates gastric cancer cell invasion under hypoxia by suppressing reactive oxygen species accumulation. Sci. Rep. 9, 2822 (2019).
Gaowa, S. et al. Possible role of p53/Mieap-regulated mitochondrial quality control as a tumor suppressor in human breast cancer. Cancer Sci. 109, 3910–3920 (2018).
Sano, H. et al. p53/Mieap-regulated mitochondrial quality control plays an important role as a tumor suppressor in gastric and esophageal cancers. Biochem. Biophys. Res. Commun. 529, 582–589 (2020).
Dan, X. et al. DNA damage invokes mitophagy through a pathway involving Spata18. Nucleic Acids Res. 48, 6611–6623 (2020).
Helleday, T., Petermann, E., Lundin, C., Hodgson, B. & Sharma, R. A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 8, 193–204 (2008).
Towers, C. G. et al. Mitochondrial-derived vesicles compensate for loss of LC3-mediated mitophagy. Dev. Cell 56, 2029–2042.e2025 (2021). This study demonstrates that when macromitophagy is blocked, autophagy-dependent cancer cells can utilize MDVs to degrade damaged mitochondria for survival.
Noguchi, S. & Shimizu, S. Molecular mechanisms and biological roles of GOMED. FEBS J. https://doi.org/10.1111/febs.16281 (2021).
Tan, H. W. S. et al. A degradative to secretory autophagy switch mediates mitochondria clearance in the absence of the mATG8-conjugation machinery. Nat. Commun. 13, 3720 (2022).
König, T. et al. MIROs and DRP1 drive mitochondrial-derived vesicle biogenesis and promote quality control. Nat. Cell Biol. https://doi.org/10.1038/s41556-021-00798-4 (2021). This study shows that the formation of TOMM20-positive MDVs is mediated by MIRO1, MIRO2 and their binding partners to form membrane protrusions first, and then DNM1L catalyses the scission of the mitochondrial protrusion to form MDVs.
Ryan, T. A. et al. Tollip coordinates Parkin-dependent trafficking of mitochondrial-derived vesicles. EMBO J. 39, e102539 (2020).
Sugiura, A., McLelland, G. L., Fon, E. A. & McBride, H. M. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33, 2142–2156 (2014).
Picca, A. et al. Generation and release of mitochondrial-derived vesicles in health, aging and disease. J. Clin. Med. https://doi.org/10.3390/jcm9051440 (2020).
Soubannier, V. et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22, 135–141 (2012). This study reveals that MDVs serve as a novel vesicle transport route to deliver mitochondrial cargoes to lysosomes, which is independent of ATG5, LC3 and mitochondrial depolarization.
Hughes, A. L., Hughes, C. E., Henderson, K. A., Yazvenko, N. & Gottschling, D. E. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. eLife https://doi.org/10.7554/eLife.13943 (2016).
English, A. M. et al. ER-mitochondria contacts promote mitochondrial-derived compartment biogenesis. J. Cell Biol. https://doi.org/10.1083/jcb.202002144 (2020).
Kanki, T. et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 14, 788–794 (2013).
Kanki, T., Wang, K., Cao, Y., Baba, M. & Klionsky, D. J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 (2009).
Li, B. et al. Mitochondrial-derived vesicles protect cardiomyocytes against hypoxic damage. Front. Cell Dev. Biol. 8, 214 (2020).
Matheoud, D. et al. Parkinson’s disease-related proteins PINK1 and parkin repress mitochondrial antigen presentation. Cell 166, 314–327 (2016).
Henne, W. M., Stenmark, H. & Emr, S. D. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a016766 (2013).
Hurley, J. H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 20, 4–11 (2008).
Stringer, D. K. & Piper, R. C. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J. Cell Biol. 192, 229–242 (2011).
Piper, R. C., Dikic, I. & Lukacs, G. L. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a016808 (2014).
Denzer, K., Kleijmeer, M. J., Heijnen, H. F., Stoorvogel, W. & Geuze, H. J. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113, 3365–3374 (2000).
Ganesan, D. & Cai, Q. Understanding amphisomes. Biochem. J. 478, 1959–1976 (2021).
Leibiger, C. et al. TDP-43 controls lysosomal pathways thereby determining its own clearance and cytotoxicity. Hum. Mol. Genet. 27, 1593–1607 (2018).
Li, J. & Hochstrasser, M. Microautophagy regulates proteasome homeostasis. Curr. Genet. https://doi.org/10.1007/s00294-020-01059-x (2020).
Segev, N. ESCRTing proteasomes to the lysosome. PLoS Genet. 16, e1008631 (2020).
Iwama, R. & Ohsumi, Y. Analysis of autophagy activated during changes in carbon source availability in yeast cells. J. Biol. Chem. 294, 5590–5603 (2019).
Krüger, U., Wang, Y., Kumar, S. & Mandelkow, E. M. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol. Aging 33, 2291–2305 (2012).
Schaeffer, V. et al. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy. Brain 135, 2169–2177 (2012).
Lu, K., Psakhye, I. & Jentsch, S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158, 549–563 (2014).
Lin, C. Y. et al. Autophagy receptor tollip facilitates bacterial autophagy by recruiting galectin-7 in response to group A Streptococcus infection. Front. Cell Infect. Microbiol. 10, 583137 (2020).
Acknowledgements
The authors apologize to colleagues whose work could not be cited owing to space limitations. This work was supported by the following grants: NMRC/CIRG/1490/2018 from the Singapore National Medical Research Council, MOE2018-T2-1-060 from the Singapore Ministry of Education, SRG2020-00002-FHS CPG2020-00029-FHS and CPG2021-00004-FHS from the University of Macau and FDCT0078/2020/A2 and 0031/2021/A1 from the Macau Science and Technology Development Fund to H.M.S., NIH grant GM131919 to D.J.K. and grant number 82071441 from the National Natural Science Foundation of China to L.M.W.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Patrice Codogno, Suresh Subramani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Multivesicular body
-
(MVB). A late endosome formed by invagination of the limiting membrane. On the one hand, MVBs can fuse with an autophagosome to form an amphisome in macroautophagy; on the other hand, in microautophagy, MVBs contain hydrolytic enzymes that function similarly to lysosomes or fuse with lysosomes to degrade autophagic cargoes.
- Soluble N-ethylmaleimide-sensitive factor attachment protein receptor
-
(SNARE). A complex consisting of the SNARE proteins, including Qa, Qb, Qc and R fusion proteins. SNARE complex assembly, through a hetero-oligomeric or four-helix bundle, is responsible for the fusion of different membranes.
- ESCRTs
-
The core ESCRTs machinery consists of five distinct protein complexes — ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III and vacuolar protein sorting-associated protein 4 (VPS4) — which can mediate different membrane fission events. ESCRTs machinery is also involved in protein sorting and endosomal intralumenal vesicle formation.
- ER-associated degradation
-
A protein quality control system to degrade misfolded proteins of the endoplasmic reticulum (ER). These misfolded proteins are retrotranslocated through the ER membrane and are ubiquitylated by a membrane-associated ubiquitin E3 ligase complex. After ubiquitylation, these substrates are extracted from the ER membrane in an ATP-dependent manner and released to the cytosol for degradation by the proteasome.
- ER whorls
-
During endoplasmic reticulum (ER) stress, subdomains of the ER are stacked together to form multilamellar membrane whorl structures, which might be mediated by eukaryotic translation initiation factor 2α kinase 3 (EIF2AK3) and the coat protein complex II (COPII) machinery in mammalian cells.
- LC3
-
(MAP1LC3). LC3 is a homologue of yeast Atg8. Cytosolic LC3-I can be conjugated to phosphatidylethanolamine to become LC3-II, which is frequently used as an autophagic marker. The LC3 family includes LC3A, LC3B, LC3B2 and LC3C, all of which are involved in the biogenesis of autophagosomes, in cargo recruitment and in the fusion of autophagosomes with lysosomes.
- ER exit site
-
(ERES). A subdomain of the endoplasmic reticulum (ER) where the coat protein complex II (COPII)-positive buds are formed.
- Translocon
-
Protein complex involved in the transport of polypeptides and/or proteins across membranes. On the endoplasmic reticulum membrane, SEC61 subunits including SEC61A, SEC61B and SEC61G form a core channel associated with different accessory factors, such as SEC62 and SEC63, through which proteins translocate through the endoplasmic reticulum membrane.
- LC3-interating region
-
(LIR). WXXL-like sequences in proteins such as sequestosome 1 (SQSTM1) and BCL-2-interacting protein 3-like protein (BNIP3L) that mediate binding to ATG8-family proteins.
- Micronuclei
-
Small isolated nuclei that contain chromosomes and/or their fragments within the nuclear envelope, which are not incorporated into the daughter nucleus after cell division and are susceptible to rupture and degradation.
- Homotypic fusion and protein-sorting complex
-
(HOPS complex). A protein complex consisting of vacuolar protein sorting-associated protein 11 (VPS11), VPS16, VPS18, VPS33A, VPS39 and VPS41 that mediates the fusion of late endosomes with lysosomes.
- LC3-associated phagocytosis
-
Phagocytosis in macrophages that needs the conjugation of microtubule-associated protein 1 light chain 3 (MAP1LC3; also known as LC3)-family proteins to the single-membrane phagosome to promote phagosome acidification and fusion with lysosomes. This process is involved in immune regulation and inflammatory responses.
- UNC-51-like autophagy-activating kinase 1
-
(ULK1). A mammalian homologue of yeast Atg1 which is required for macroautophagy to initiate the formation of the phagophore, and to recruit and release other autophagy-related proteins from the phagophore assembly site.
- Intralumenal vesicles
-
Vesicles that are formed through invagination of the endosomal membrane or another membrane and hence reside within the lumen of the corresponding compartment.
- ATG5
-
The gene encoding the protein ATG5, which is member of the ATG12–ATG5–ATG16L1 complex, which functions partly as an E3 ligase for ATG8-family protein conjugation to phosphatidylethanolamine.
- Kufor-Rakeb syndrome
-
A rare form of inherited juvenile-onset atypical Parkinson disease, also known as Parkinson disease 9. Individuals with Kufor-Rakeb syndrome usually start to develop symptoms between 10 years and 20 years of age. In addition to the typical Parkinson disease symptoms, patients with Kufor-Rakeb syndrome can also experience atypical symptoms, such as supranuclear gaze palsy, paraplegia and ataxia.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, L., Klionsky, D.J. & Shen, HM. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol 24, 186–203 (2023). https://doi.org/10.1038/s41580-022-00529-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41580-022-00529-z
- Springer Nature Limited
This article is cited by
-
Mitochondrial protein C15ORF48 is a stress-independent inducer of autophagy that regulates oxidative stress and autoimmunity
Nature Communications (2024)
-
OPN silencing reduces hypoxic pulmonary hypertension via PI3K-AKT-induced protective autophagy
Scientific Reports (2024)
-
Ajugol's upregulation of TFEB-mediated autophagy alleviates endoplasmic reticulum stress in chondrocytes and retards osteoarthritis progression in a mouse model
Chinese Medicine (2023)
-
Phospholipase A2 regulates autophagy in gouty arthritis: proteomic and metabolomic studies
Journal of Translational Medicine (2023)
-
Autophagy in BRAF-mutant cutaneous melanoma: recent advances and therapeutic perspective
Cell Death Discovery (2023)