Abstract
This review focuses on the structure and mode-of-action of the two-peptide (class-IIb) bacteriocins that consist of two different peptides whose genes are next to each other in the same operon. Optimal antibacterial activity requires the presence of both peptides in about equal amounts. The two peptides are synthesized as preforms that contain a 15–30 residue double-glycine-type N-terminal leader sequence that is cleaved off at the C-terminal side of two glycine residues by a dedicated ABC-transporter that concomitantly transfers the bacteriocin peptides across cell membranes. Two-peptide bacteriocins render the membrane of sensitive bacteria permeable to a selected group of ions, indicating that the bacteriocins form or induce the formation of pores that display specificity with respect to the transport of molecules. Based on structure–function studies, it has been proposed that the two peptides of two-peptide bacteriocins form a membrane-penetrating helix–helix structure involving helix–helix-interacting GxxxG-motifs that are present in all characterized two-peptide bacteriocins. It has also been suggested that the membrane-penetrating helix–helix structure interacts with an integrated membrane protein, thereby triggering a conformational alteration in the protein, which in turn causes membrane-leakage. This proposed mode-of-action is similar to the mode-of-action of the pediocin-like (class-IIa) bacteriocins and lactococcin A (a class-IId bacteriocin), which bind to a membrane-embedded part of the mannose phosphotransferase permease in a manner that causes membrane-leakage and cell death.
Similar content being viewed by others
Introduction
Bacteria produce ribosomally synthesized antibacterial polypeptides, generally referred to as bacteriocins [12, 20, 24, 31, 52, 53, 55]. These polypeptides have attracted much attention due to their potential use as antibacterial agents for the treatment of infections and preservation of food and animal feed. There has especially been great interest in the peptide bacteriocins produced by lactic acid bacteria (LAB) because of the “food grade quality” and industrial importance of these bacteria. LAB are used in food and feed production, they are part of the natural microbial flora in food humans have consumed for centuries, and they constitute a significant part of the indigenous microbiota of mammals, including humans. It is therefore considered to be relatively safe to use these bacteria and the bacteriocins they produce to prevent the growth of pathogenic/undesirable bacteria [9]. The LAB bacteriocins pediocin PA-1/AcH and nisin are in fact presently used as food preservatives [11, 12, 15], and the potential of LAB bacteriocins in medical applications is illustrated by the fact that oral intake of bacteriocin-producing LAB protects mice against lethal doses of Listeria monocytogenes [10].
The small heat stable LAB peptide bacteriocins are divided into two classes.Footnote 1 Class-I consists of bacteriocins (often referred to as lantibiotics) that contain one or more of the modified amino acid residues lanthionine, β-methyllanthionine, dehydroalanine, dehydrobutyrine, and/or d-alanine [11, 12], whereas class-II consists of bacteriocins that lack modified residues [12, 20, 24, 53, 55]. The class-II bacteriocins are further divided into four subclasses, class-IIa, -IIb, -IIc, and -IId [12]. Class-IIa contains the antilisterial one-peptide pediocin-like bacteriocins that have similar amino acid sequences [12, 20, 24, 53], class-IIb contains the two-peptide bacteriocins [12, 31, 53, 55], class-IIc consists of the cyclic bacteriocins whose N- and C-termini are covalently linked [12, 53], and class-IId contains the one-peptide non-cyclic bacteriocins that show no sequence similarity to the pediocin-like bacteriocins [12, 53].
Developing LAB bacteriocins into agents for treatment of infections and for preservation of food and animal feed is clearly of considerable importance due to the alarming increase in antibiotic-resistant pathogenic bacteria and the undesirable side effects that many chemical preservatives may have. Rational design of new bacteriocin variants with properties that make them especially useful for medical and biotechnological applications requires, however, insight into how they function at a molecular level. This in turn necessitates insight into their three-dimensional structures, as bacteriocins function through structural interactions. In this review on the two-peptide (class-IIb) bacteriocins, the focus will especially be on recent structure–function studies that have given us some insight into the structure of these bacteriocins.
Proteins and Genes Involved in Production of Two-Peptide Bacteriocins
At least 15 two-peptide (class-IIb) bacteriocinsFootnote 2 (Fig. 1) have been isolated and characterized since the first isolation of such a bacteriocin (lactococcin G) in 1992 [51]. As the name suggests, these bacteriocins are unique in that they consist of two different peptides, and optimal antibacterial activity requires the presence of both peptides in about equal amounts [31, 53, 55]. The two peptides are synthesized as preforms that contain a 15–30 residue N-terminal leader sequence of the so-called double-glycine type that is cleaved off at the C-terminal side of two glycine residues by a dedicated ABC-transporter that transfers the bacteriocin across the membrane [37, 38]. The leader sequence apparently facilitates interaction with the dedicated ABC-transporter and might possibly also function to keep the bacteriocin inactive until it has been secreted. The genes encoding the preforms of the two peptides are always found next to each other in the same operon along with the gene that encodes the immunity protein that protects the bacteriocin-producer from being killed by its own bacteriocin [53, 55]. For some bacteriocins (such as lactococcin G), this operon contains also the genes encoding the dedicated ABC-transporter and a so-called accessory protein. For other bacteriocins (such as enterocin 1071, plantaricin E/F, and plantaricin J/K), the genes encoding the ABC-transporter and accessory protein are on a separate operon nearby the operon with the genes encoding the preforms of the two peptides and the immunity protein. The function of the so-called accessory protein is not entirely clarified, but it may be involved in immunity against the bacteriocin and/or required (along with the dedicated ABC-transporter) for secretion of the bacteriocin.
Amino acid sequences of the unmodified (class-IIb) two-peptide bacteriocins. The GxxxG-motifs are marked with black background. Plantaricin Sβ and plantaricin NC8β have the GxxxG-like motifs AxxxA- and SxxxS-motif, respectively, instead of the GxxxG-motif. The recently identified two-peptide bacteriocin enterocin C [43] is not included in the figure. Its sequence is identical to enterocin 1071, except that enterocin C has an alanine residue instead of a threonine residue at position 17 in its β-peptide [43]. The two-peptide bacteriocins leucocin H [7], lactococcin MN [65], and lactococcin MMT24 [32] are also not included in the figure. Leucocin H has not been completely sequenced, but the partial sequence reveals a putative GxxxG-motif. Also lactococcin MN has putative GxxxG-motifs, but is not included in the list, since only the sequences of the preforms are known, and the cleavage site is unknown. Lactococcin MMT24 has not been sequenced. The figure is a modified version of Fig. 1 in Ref. [56]. References for the sequences are indicated in the figure, and are as follows: lactococcin G [51], lactococcin Q [67], enterocin 1071 [4, 5, 27], plantaricin E/F and J/K [3, 17], plantaricin S [39, 63], plantaricin NC8 [45], lactacin F [2, 29], brochocin C [47], thermophilin 13 [46], ABP-118 [26], salivaricin P (differs from ABP-118 in only two residues) [6], mutacin IV [57], and lactocin 705 [14]
Some two-peptide bacteriocins seem to be produced constitutively, whereas the production of other two-peptide bacteriocins (such as plantaricin E/F, plantaricin J/K [16–18], ABP-118 [26] and NC8 [44]) is transcriptionally regulated through a three-component regulatory system consisting of a peptide pheromone, a membrane-associated histidine protein kinase, and response regulators [41]. Upon secretion from bacteria, the peptide pheromone interacts with the membrane-associated histidine kinase, thereby triggering the kinase to phosphorylate the intracellular response regulator, thus enabling the response regulators to activate the operon needed for bacteriocin synthesis and secretion [41].
Plantaricin A is the peptide pheromone that induces the production of two two-peptide bacteriocins, plantaricin E/F and plantaricin J/K, from Lactobacillus plantarum C11 [16–18, 34]. Structural analysis of this peptide pheromone revealed a novel membrane-interacting mechanism whereby the pheromone (and perhaps other membrane active peptide pheromones/hormones) functions [42]. It seems that plantaricin A first interacts in a non-chiral manner with membrane lipids, and that this interaction induces α-helical structuring in a region of the peptide [42]. The pheromone thereby becomes sufficiently structured and properly positioned in the membrane to subsequently interact in a chiral manner with the histidine protein kinase in or near the membrane–water interface.
The Two Peptides of Two-Peptide Bacteriocins Function as One Antibacterial Unit
A peptide from a two-peptide bacteriocin displays high antimicrobial activity only when combined with the complementary peptide from the same two-peptide bacteriocin or in some cases when combined with a peptide from a homologous two-peptide bacteriocin. For instance, the two peptides that constitute lactococcin G are active at pico to nano-molar concentrations when combined, but show no activity when assayed individually at micro-molar concentrations [49, 51], nor when combined with either the E- or F-peptide of plantaricin E/F or the J- or K-peptide of plantaricin J/K [3]. High activity is, however, obtained when one of the lactococcin G peptides is combined with the complementary peptide from lactococcin Q or enterocin 1071 [54, 55, 67], presumably, because these three two-peptide bacteriocins show extensive sequence similarities (about 88 and 57% sequence identity between lactococcin G and, respectively, lactococcin Q and enterocin 1071; Fig. 1).
The fact that a peptide from a two-peptide bacteriocin displays high antibacterial activity only in combination with the complementary peptide from the same or a homologous two-peptide bacteriocin clearly indicates that the two peptides of two-peptide bacteriocins function together as one antibacterial unit. This is in accordance with results showing that the genes encoding the two peptides of two-peptide bacteriocins are always next to each other on the same operon, and that the two peptides are thus produced in approximately equal amounts. Moreover, circular dichroism (CD) structural studies indicate that the two peptides of two-peptide bacteriocins interact and structure each other when exposed simultaneously to membrane-like entities such as liposomes [33, 35]. The synergistic action of the two peptides thus seems to be due to the fact that they interact with each other and form one antibacterial unit, rather than that they separately act on two different sites on target cells.
The Two-Peptide Bacteriocins Permeabilize Target-Cell Membranes
All two-peptide bacteriocins whose mode-of-action has been studied, this includes plantaricin E/F [50], plantaricin J/K [50], lactococcin G [48, 49], thermophilin 13 [46], lactacin F [1], and lactocin 705 [8, 13], render the membranes of sensitive bacteria permeable to small molecules. The studies revealed that the bacteriocins show specificity with respect to which small molecules they conduct across membranes, and that the bacteriocins differ somewhat in their specificities. For instance, lactococcin G permeabilizes target-cell membranes for a variety of monovalent cations, such as Na+, K+, Li+, Cs+, Rb+, and choline, but not for H+, nor divalent cations such as Mg2+ or anions such as phosphate [48, 49]. Plantaricin E/F and plantaricin J/K permeabilize membranes for monovalent ions, including H+ (in contrast to lactococcin G), but not for divalent ions such as phosphate and Mg2+ [50]. It appears, however, as if plantaricin J/K conducts anions more efficiently than plantaricin E/F and vice versa for cations [50]. Lactacin F renders membranes permeable to K+ and phosphate [1], and thermophilin 13 has been shown to dissipate both the trans-membrane electrical potential and pH gradient, but its specificity with respect to molecules it conducts across membranes has not been studied [46]. This ability to differentiate between molecules they transport across membranes indicates that these two-peptide bacteriocins form (or induce the formation of) relatively sophisticated pores that display specificity with respect to transport of molecules, rather than induce membrane-leakage through a detergent-like disruption of membranes. Moreover, the high potency of the two-peptide bacteriocins suggests that membrane permeabilization depends on a relative low number of peptides, unlike what is expected if permeabilization is due to a detergent-like disruption of membranes.
The Structure of Two-Peptide Bacteriocins
CD and NMR structural studies have been carried out on the three two-peptide bacteriocins lactococcin G, plantaricin E/F, and plantaricin J/K [25, 33, 35, 58, 59]. CD studies show that the peptides of these three bacteriocins are unstructured, with no structural interactions between complementary peptides, when in aqueous solution. Helical structuring occurs, however, when the peptides are individually exposed to a more hydrophobic or membrane-like environment, such as when the peptides are in solutions containing trifluoroethanol (TFE), micelles or liposomes [25, 33, 35, 58, 59]. Moreover, additional structuring is obtained when complementary peptides are mixed and then exposed to membrane-like entities such as liposomes, indicating that the peptides interact in a structure-inducing manner upon contact with target-membranes [33, 35]. It seems that the target-membrane must have a reasonable flat surface (i.e., liposomes or bilayers) for the inter-peptide interaction to take place, since the additional structuring is not observed in the presence of micelles.
All two-peptide bacteriocins identified so far contain GxxxG-motifs (Fig. 1). This motif is significantly over-represented in transmembrane helices and it is, along with the “GxxxG-like” motifs AxxxA and SxxxS, known to mediate helix–helix interactions in membrane proteins [60–62]. When in an α-helix, the two glycine residues in the motif will be on the same side of the helix and thereby create a flat interaction site that allows close inter-helical contact, thereby enabling optimal inter-helical van der Waals interactions and formation of stabilizing inter-helical backbone Cα–H…O hydrogen bonds [62].
The presence of GxxxG-motifs and a high helical content suggests that the peptides of two-peptide bacteriocins form membrane-penetrating helix–helix structures involving their GxxxG-motifs. Such helix–helix structuring would generate and/or stabilize helical structuring in the somewhat flexible GxxxG-motifs and possibly also in other flexible segments in the two complementary peptides, and this could explain the increased helical content observed by CD spectroscopy when complementary peptides are simultaneously exposed to target-membranes.
Structure and Membrane-Orientation of Lactococcin G
Lactococcin G is perhaps the two-peptide bacteriocin that is best characterized [35, 48, 49, 51, 54, 56, 58]. It consists of the 39 residue α-peptide (often termed LcnG-α) and the 35 residue β-peptide (often termed LcnG-β) (Fig. 1). NMR structural studies revealed α-helices in both the N- (residues 3–21) and C- (residues 24–34) terminal halves of the α-peptide in the presence of either DPC micelles or TFE [58]. Helices were also found in the N- (residues 11–19) and C- (residues 23–32) terminal halves of the β-peptide in the presence of TFE, but only one helix, in the N-terminal half, was observed in the presence of DPC [58].
The α-peptide has two GxxxG-motifs (Gly-7 to Gly-11 and Gly-18 to Gly-22; Fig. 1), and the β-peptide has one (Gly-18 to Gly-22; Fig. 1). The GxxxG-motif in the β-peptide and the first one (G7xxxG11) in the α-peptide are also found in lactococcin Q and enterocin 1071 (Fig. 1), two bacteriocins that have extensive sequence similarity with lactococcin G. This suggests that these two GxxxG-motifs are of special importance for the functioning of lactococcin G, lacococcin Q, and enterocin 1071, and it has been postulated that the α- and β-peptides of these three bacteriocins interact through helix–helix interactions involving these two GxxxG-motifs. Indeed, replacement of the glycine residues in these GxxxG-motifs with larger residues that interfere with close helix–helix contact was highly detrimental, consistent with these motifs being involved in helix–helix interactions [56]. Replacement of the other glycine residues in the peptides was, in contrast, relatively well tolerated [56].
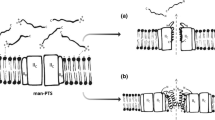
Based on site-directed mutagenesis studies (discussed in the three subsequent paragraphs) and the NMR structure of the α- and β-peptides, a structural model of lactococcin G has been proposed [53, 56, 58]. The proposed structure should also be valid for enterocin 1071 and lactococcin Q, since their sequence similarities to lactococcin G indicate that these three bacteriocins have similar three-dimensional structures and mode-of-action. The proposed structure entails that the α- and β-peptides are oriented in the same direction and form a staggered helix–helix structure that spans the target-cell membrane (Fig. 2) [53, 56, 58]. The helix–helix segment consists of the N-terminal half of the α-peptide (from about residue 3 to residue 22) and the C-terminal half of the β-peptide (from about residue 13 to residue 32). The structural model also entails that the strikingly cationic C-terminal end (residues 35–39: R–K–K–K–H) of the α-peptide is unstructured and forced through the target-cell membrane by the membrane potential (negative inside), thereby positioning the C-termini of the two peptides inside the target-cell (Fig. 2). The tryptophan-rich N-terminal end of the β-peptide is also proposed to be relatively unstructured and to position itself in the outer membrane interface, thus forcing the N-termini of the two peptides to remain on the outer side of the target-cell membrane and the helix–helix segment to transverse the membrane (Fig. 2). The proposed helix–helix interaction will structure the flexible G18xxxG22-motif in the β-peptide and thereby prolong the main helix from residue 19 to at least residue 22. The helix–helix interaction may also generate and/or stabilize helical structuring in other regions as well, such as between residues 21 and 24 (which is part of the second GxxxG-motif) in the α-peptide and thereby prolong the main helix in the α-peptide to residue 34. This may explain the increased helicity observed by CD spectroscopy when complementary peptides are mixed together and exposed to target-membranes [33, 35].
Cartoon representation of the structural model of lactococcin G and its orientation in target-cell membranes. The two peptides interact through the G7xxxG11-motif in the α-peptide and the G18xxxG22-motif in the β-peptide and form a trans-membrane helix–helix structure. The highly positively charged and structurally flexible C-terminal end of the α-peptide is forced through the membrane by the trans-membrane potential (negative inside). The tryptophan residues in the structurally flexible N-terminal region of the β-peptide are in or near the outer membrane interface
The four C-terminal residues in the α-peptide and four of the five N-terminal residues in the β-peptide can be replaced with the corresponding D-isomeric residues without a high reduction in the antibacterial activity (Oppegård, Rogne, unpublished results). The C- and N-terminal region of, respectively, the α- and β-peptide are thus not highly structured nor involved in inter-peptide interactions—consistent with the proposed structure. Also the replacement of any one of the three tryptophan residues (Trp-3, Trp-5 and Trp-8) in the structurally flexible N-terminal region of the β-peptide with an aromatic (Phe, Tyr), hydrophobic (Leu), or hydrophilic (Arg) residue is relatively well tolerated, indicating that this region is not embedded in a strictly hydrophobic or hydrophilic environment [56]. In accordance with the proposed structure, this suggests that the region is in fact embedded in the membrane interface, thus enabling the side chains of the hydrophilic and hydrophobic residues to “snorkel” into the, respectively, hydrophilic and hydrophobic regions of the membrane.
Also consistent with the proposed structure (which entails that the N-termini of the peptides face the cells outside) are results showing that fusion proteins consisting of the immunoglobulin-binding domain of streptococcal protein G linked to the N-terminal residue of either the α- or β-peptide exhibit antibacterial activity in combination with the complementary peptide. Although the activity was reduced dramatically (about 20,000-fold reduction for the fused α-peptide and 5,000-fold reduction for the fused β-peptide), the results suggest that the N-terminal part of the peptides does not penetrate through the target-cell membrane (P. Rogne and C. Oppegård, unpublished results).
One conspicuous difference between the homologous bacteriocins lactococcin G, lactococcin Q, and enterocin 1071 is that the latter bacteriocin has a negative charge (which is absent in lactococcin G and lactococcin Q) at the N-terminus of the α-peptide, whereas lactococcin G and lactococcin Q have a negative charge (which is absent in enterocin 1071) at position 10 in the β-peptide (Fig. 1). Site-directed mutagenesis studies has revealed that the detrimental effect of removing the N-terminal negative charge in the α-peptide of enterocin 1071 is neutralized by introducing a negative charge at position 10 in the β-peptide of enterocin 1071 [54]. The negative charge in residue-1 in the α-peptide (as in enterocin 1071) thus appears to be functionally equivalent to a negative charge in residue-10 in the β-peptide (as in lactococcin G and lactococcin Q). The proposed structure may account for this, since residue-1 in the α-peptide and residue-10 in the β-peptide are near each other in the proposed structure (Fig. 2). The proposed structure may also explain mutagenesis results showing that the lactococcin G immunity protein recognizes both the N-terminal region (residues 1–13) of the α-peptide and a sequence on the C-terminal side of residue 13 in the β-peptide (C. Oppegård et al., in preparation), since these regions are adjacent to each other in the proposed structure (Fig. 2) and may consequently interact simultaneously with the immunity protein.
Structure of Plantaricin E/F
The three-dimensional structures of the E (33 residues) and F (34 residues) peptides that constitute plantaricin E/F have also been analyzed by NMR spectroscopy [25]. In the presence of DPC micelles, the E-peptide forms two α-helical-like regions (residues 10–21 and 25–31) separated by a flexible GxxxG-motif (residues 20–24), whereas the F-peptide forms one long helix from residue 7 to 32, with a kink and slightly more flexible region around Pro-20 [25]. It was proposed that the two peptides interact and form a helix–helix structure involving their GxxxG-motifs [25]. The E-peptide has two such motifs, G5xxxG9 and G20xxxG24, while the F-peptide has one, G30xxxG34 (Fig. 1).
It is, however, not clear how these GxxxG-motifs might combine, and several alternatives have been suggested [25]. One possibility is that the G30xxxG34-motif in the F-peptide pairs with the G5xxxG9-motif in the E-peptide. This pairing is not so likely, however, since it would result in a very short (not more than 9 residues in length) helix–helix region if the motifs interacted in a parallel orientation, and the negatively charged aspartate residues in the E- and F-peptides (D17 and D22 in the E- and F-peptides, respectively) would come near each other if they interacted in an antiparallel orientation [25]. A more likely pairing is that G20xxxG24 in the E-peptide interacts in a parallel orientation with G30xxxG34 in the F-peptide, as this will bring G5xxxG9 in the E-peptide near a flat surface formed by residues S16, G19, and P20 in the F-peptide [25]. Another possibility is that the flat surface formed by residues S16, G19, and P20 in the F-peptide interacts with G20xxxG24 in the E-peptide. Future mutagenesis analysis of the E- and F-peptides should differentiate between these alternatives.
Structure of Plantaricin J/K
The three-dimensional structures of the J- (25 residues) and K- (32 residues) peptides that constitute plantaricin J/K were recently also analyzed by NMR spectroscopy [59]. In the presence of membrane-mimicking environments (DPC micelles or TFE), the J-peptide forms an N-terminal amphiphilic α-helix between residues 3 and 15, while the K-peptide forms a central amphiphilic α-helix between residues 9 and 24. There is one GxxxG-motif (Gly-13 to Gly-17) in the J-peptide and two (Gly-9 to Gly-13 and Gly-13 to Gly-17) in the K-peptide (Fig. 1). Testing the activity of single glycine replacement variants of the two peptides revealed that replacing glycine residues that are part of the G13xxxG17-motifs in the J- and K-peptides with large residues was detrimental, while similar replacements of the other glycine residues in these two peptides were relatively well tolerated [59]. These mutagenesis results combined with the NMR structures of the J- and K-peptides are in accordance with a structural model in which the two peptides interact and form a helix–helix structure involving their G13xxxG17-motifs [59]. It was, however, not possible to deduce which of the two possible peptide orientations, the parallel or the antiparallel, is the most plausible orientation [59].
Some Bacteriocins Function as Peptide Ligands that Interact with Membrane-Associated Receptors
Mode-of-action studies on the pediocin-like (class-IIa) bacteriocins and the one-peptide non-pediocin-like (i.e., class-IId) bacteriocin lactococcin A revealed that these bacteriocins bind to a part of the mannose phosphotransferase permease (the MptC and/or MptD subunits) that is embedded in the cell membrane [19]. The study also revealed that the immunity proteins that protect cells from these bacteriocins bind strongly to the bacteriocin–permease complex and thereby prevent bacteriocin-induced killing [19]. Interactions between these bacteriocins and the mannose phosphotransferase permease thus apparently alter the conformation of the permease in a manner that results in membrane-leakage, and this leakage may be blocked on the cytosolic side of the membrane by the binding of an immunity protein to the bacteriocin–permease complex.
Structure–function analyses of the pediocin-like (class-IIa) bacteriocins revealed that these bacteriocins contain a helical region that penetrates into the target-membrane and is involved in determining the antimicrobial spectrum [21, 22, 24, 28, 36, 40, 64, 66]. The membrane-penetrating helix may be the structural motif that interacts with the mannose phosphotransferase system, as 15-mer fragments covering most of the helix inhibit these bacteriocins in a specific manner [23; Haugen et al., to be submitted]. It has indeed been suggested that helical interactions between peptide bacteriocins and integrated membrane (transport) proteins might be a common mechanism whereby peptide bacteriocins cause membrane-leakage [53, 56, 58]. In the case of two-peptide bacteriocins, this interaction might involve the trans-membrane helix–helix structure. Such a structure might interact with an integrated membrane protein in an analogous fashion as shown to be the case for lactococcin A and the pediocin-like (class-IIa) bacteriocins [19], although a membrane-associated receptor for a two-peptide bacteriocin has yet to be identified. It has, however, been speculated that the lactococcin G immunity protein interacts with lactococcin G indirectly via a putative lactococcin G receptor in an analogous manner as discussed for the pediocin-like immunity proteins. The functionality of the lactococcin G immunity protein is indeed target-cell dependent (as is the case for the pediocin-like immunity proteins) and thus requires a cellular component (C. Oppegård, in preparation), and this cellular component could perhaps be a receptor for lactococcin G.
Although five two-peptide bacteriocinsFootnote 3 adopt mainly helical structures, it should be mentioned that this may not be the case for all two-peptide bacteriocins. CD and NMR analyses of the two-peptide bacteriocin brochocin-C suggest a high content of β-sheet structure [30], and this may possibly also be the case for thermophilin 13, since it has marked sequence similarity with brochocin-C [46, 47]. However, also these two bacteriocins contain several GxxxG-motifs that suggest helix–helix interactions between their complementary peptides. The structures of brochocin-C and thermophilin 13 must be analyzed in more detail before it can be concluded whether or not they in fact have a high content of β-sheet structure.
It is essential that more extensive structural analyses of two-peptide bacteriocins (and other peptide bacteriocins) are performed, as this is necessary to attain more certain and detailed insight into the three-dimensional structure of these bacteriocins, which in turn should reveal structural features that are important for their function and potency. Such insight will be invaluable for future rational design of bacteriocin variants with properties that make them useful for medical and biotechnological applications, and may allow the development of these peptide bacteriocins into additives for treatment of infections and preservation of food and animal feed.
Notes
There is also a third class of LAB bacteriocins (class III), that consists of a few large heat-labile protein bacteriocins that have overall not been as extensively characterized as the class I and II peptide bacteriocins.
Lactococcin G, plantaricin E/F and plantaricin J/K, as judged by NMR analyses, and lactococcin Q and enterocin 1071, as deduced from their similarity to lactococcin G.
References
Abee T, Klaenhammer TR, Letellier L (1994) Kinetic studies of the action of lactacin F, a bacteriocin produced by Lactobacillus johnsonii that forms poration comlexes in the cytoplasmic membrane. Appl Environ Microbiol 60:1006–1013
Allison GE, Fremaux C, Klaenhammer TR (1994) Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J Bacteriol 176:2235–2241
Anderssen EL, Diep DB, Nes IF, Eijsink VGH, Nissen-Meyer J (1998) Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricin EF and JK, and the induction factor, plantaricin A. Appl Environ Microbiol 64:2269–2272
Balla E, Dicks LMT (2005) Molecular analysis of the gene cluster involved in the production and secretion of enterocins 1071A and 1071B and of the genes responsible for the replication and transfer of plasmid pEF1071. Int J Food Microbiol 99:33–45
Balla E, Dicks LMT, Du Toit M, van der Merwe MJ, Holzapfel WH (2000) Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Appl Environ Microbiol 66:1298–1304
Barrett E, Hayes M, O’Connor P, Gardiner G, Fitzgerald GF, Stanton C, Ross RP, Hill C (2007) Salivaricin P, one of a family of two-component antilisterial bacteriocins produced by intestinal isolates of Lactobacillus salivarius. Appl Environ Microbiol 73:3719–3723
Blom H, Katla T, Holck A, Sletten K, Axelsson L, Holo H (1999) Characterization, production, and purification of leucocin H, a two-peptide bacteriocin from Leuconostoc MF215B. Curr Microbiol 39:43–48
Castellano P, Raya R, Vignolo G (2003) Mode of action of lactocin 705, a two-component bacteriocin from Lactobacillus casei CRL705. Int J Food Microbiol 85:35–43
Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71:1–20
Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CGM (2007) Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci 104:7617–7621
Cotter PD, Hill C, Ross RP (2005) Bacterial lantibiotics: strategies to improve therapeutic potential. Curr Protein Pept Sci 6:61–75
Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788
Cuozzo SA, Castellano P, Sesma FJM, Vignolo GM, Raya RR (2003) Differential roles of the two-component peptides of lactocin 705 in antimicrobial activity. Curr Microbiol 46:180–183
Cuozzo SA, Sesma F, Palacios JM, de Ruiz Holgado AP, Raya RR (2000) Identification and nucleotide sequence of genes involved in the synthesis of lactocin 705, a two-peptide bacteriocin from Lactobacillus casei CRL 705. FEMS Microbiol Lett 185:157–161
Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J (1996) Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69:193–202
Diep DB, Håvarstein LS, Nes IF (1995) A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol Microbiol 18:631–639
Diep DB, Håvarstein LS, Nes IF (1996) Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol 178:4472–4483
Diep DB, Myhre R, Johnsborg O, Aakra Å, Nes IF (2003) Inducible bacteriocin production in Lactobacillus is regulated by differential expression of pln operons and by two antagonizing response regulators, the activity of which is enhanced upon phosphorylation. Mol Microbiol 47:483–494
Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF (2007) Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci USA 104:2384–2389
Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H (2006) The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev 70:564–582
Fimland G, Blingsmo OR, Sletten K, Jung G, Nes IF, Nissen-Meyer J (1996) New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol 62:3313–3318
Fimland G, Eijsink VGH, Nissen-Meyer J (2002) Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508–9515
Fimland G, Jack R, Jung G, Nes IF, Nissen-Meyer J (1998) The bactericidal activity of pediocin PA-1 is specifically inhibited by a 15-mer fragment that spans the bacteriocin from the center toward the C terminus. Appl Environ Microbiol 64:5057–5060
Fimland G, Johnsen L, Dalhus B, Nissen-Meyer J (2005) Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J Pept Sci 11:688–696
Fimland N, Rogne P, Fimland G, Nissen-Meyer J, Kristiansen PE (2008) Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF. Biochim Biophys Acta 1784:1711–1719
Flynn S, van Sinderen D, Thornton GM, Holo H, Nes IF, Collins JK (2002) Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148:973–984
Franz CMAP, Grube A, Herrmann A, Abriouel H, Stärke J, Lombardi A, Tauscher B, Holzapfel WH (2002) Biochemical and genetic characterization of the two-peptide bacteriocin enterocin 1071 produced by Enterococcus faecalis FAIR-E 309. Appl Environ Microbiol 68:2550–2554
Fregeau Gallagher NL, Sailer M, Niemczura WP, Nakashima TT, Stiles ME, Vederas JC (1997) Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 36:15062–15072
Fremaux C, Ahn C, Klaenhammer TR (1993) Molecular analysis of the lactacin F operon. Appl Environ Microbiol 59:3906–3915
Garneau S, Ference CA, van Belkum MJ, Stiles ME, Vederas JC (2003) Purification and characterization of brochocin A and brochocin B(10–43), a functional fragment generated by heterologous expression in Carnobacterium piscicola. Appl Environ Microbiol 69:1352–1358
Garneau S, Martin NI, Vederas JC (2002) Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577–592
Ghrairi T, Frére J, Berjeaud JM, Manai M (2005) Lactococcin MMT24, a novel two-peptide bacteriocin produced by Lactococcus lactis isolated from rigouta cheese. Int J Food Microbiol 105:389–398
Hauge HH, Mantzilas D, Eijsink VGH, Nissen-Meyer J (1999) Membrane-mimicking entities induce structuring of the two-peptide bacteriocins plantaricin E/F and plantaricin J/K. J Bacteriol 181:740–747
Hauge HH, Mantzilas D, Moll GN, Konings WN, Driessen AJ, Eijsink VGH, Nissen-Meyer J (1998) Plantaricin A is an amphiphilic α-helical bacteriocin-like pheromone which exerts antimicrobial and pheromone activities through different mechanisms. Biochemistry 37:16026–16032
Hauge HH, Nissen-Meyer J, Nes IF, Eijsink VGH (1998) Amphiphilic α-helices are important structural motifs in the α and β peptides that constitute the bacteriocin lactococcin G: enhancement of helix formation upon α-β interaction. Eur J Biochem 251:565–572
Haugen HS, Fimland G, Nissen-Meyer J, Kristiansen PE (2005) Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide curvacin A. Biochemistry 44:16149–16157
Håvarstein LS, Diep DB, Nes IF (1995) A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol 16:229–240
Håvarstein LS, Holo H, Nes IF (1994) The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by gram-positive bacteria. Microbiology 140:2383–2389
Jiménez-Diaz R, Ruiz-Barba JL, Cathcart DP, Holo H, Nes IF, Sletten KH, Warner PJ (1995) Purification and partial amino acid sequence of plantaricin S, a bacteriocin produced by Lactobacillus plantarum LPCO10, the activity of which depends on the complementary action of two peptides. Appl Environ Microbiol 61:4459–4463
Johnsen L, Fimland G, Nissen-Meyer J (2005) The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem 280:9243–9250
Kleerebezem M, Quadri LE (2001) Peptide pheromone-dependent regulation of antimicrobial peptide production in gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579–1596
Kristiansen PE, Fimland G, Mantzilas D, Nissen-Meyer J (2005) Structure and mode of action of the membrane-permeabilizing antimicrobial peptide pheromone plantaricin A. J Biol Chem 280:22945–22950
Maldonado-Barragan A, Caballero-Guerrero B, Jimenez E, Jimenez-Diaz R, Ruiz-Barba JL, Rodriguez JM (2009) Enterocin C, a class IIb bacteriocin produced by E. faecalis C901, a strain isolated from human colostrum. Int J Food Microbiol 133:105–112
Maldonado A, Jiménez-Diaz R, Ruiz-Barba JL (2004) Induction of plantaricin production in Lactobacillus plantarum NC8 after coculture with specific gram-positive bacteria is mediated by an autoinduction mechanism. J Bacteriol 186:1556–1564
Maldonado A, Ruiz-Barba JL, Jiménez-Diaz R (2003) Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl Environ Microbiol 69:383–389
Marciset O, Jewronimus-Stratingh MC, Mollet B, Poolman B (1997) Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J Biol Chem 272:14277–14284
McCormick JK, Poon A, Sailer M, Gao Y, Roy KL, McMullen LM, Vederas JC, Stiles ME, van Belkum MJ (1998) Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl Environ Microbiol 64:4757–4766
Moll G, Hauge HH, Nissen-Meyer J, Nes IF, Konings WN, Driessen AJM (1998) Mechanistic properties of the two-component bacteriocin lactococcin G. J Bacteriol 180:96–99
Moll G, Ubbink-Kok T, Hauge HH, Nissen-Meyer J, Nes IF, Konings WN, Driessen AJM (1996) Lactococcin G is a potassium ion-conducting, two component bacteriocin. J Bacteriol 178:600–605
Moll G, van der Akker HE, Hauge HH, Nissen-Meyer J, Nes IF, Konings WN, Driessen AJM (1999) Complementary and overlapping selectivity of the two-peptide bacteriocins EF and JK. J Bacteriol 181:4848–4852
Nissen-Meyer J, Holo H, Håvarstein LS, Sletten K, Nes IF (1992) A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol 174:5686–5692
Nissen-Meyer J, Nes IF (1997) Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol 167:67–77
Nissen-Meyer J, Rogne P, Oppegård C, Haugen HS, Kristiansen PE (2009) Structure-function relationships of non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria. Curr Pharm Biotechnol 10:19–37
Oppegård C, Fimland G, Thorbæk L, Nissen-Meyer J (2007) Analysis of the two-peptide bacteriocins lactococcin G and enterocin 1071 by site-directed mutagenesis. Appl Environ Microbiol 73:2931–2938
Oppegård C, Rogne P, Emanuelsen L, Kristiansen PE, Fimland G, Nissen-Meyer J (2007) The two-peptide class II bacteriocins: structure, production, and mode of action. J Mol Microbiol Biotechnol 13:210–219
Oppegård C, Schmidt J, Kristiansen PE, Nissen-Meyer J (2008) Mutational analysis of putative helix-helix interacting GxxxG-motifs and tryptophan residues in the two-peptide bacteriocin lactococcin G. Biochemistry 47:5242–5249
Qi F, Chen P, Caufield PW (2001) The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl Environ Microbiol 67:15–21
Rogne P, Fimland G, Nissen-Meyer J, Kristiansen PE (2008) Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G. Biochim Biophys Acta 1784:543–554
Rogne P, Haugen C, Fimland G, Nissen-Meyer J, Kristiansen PE (2009) Three-dimensional structure of the two-peptide bacteriocin plantaricin JK. Peptides 30:1613–1621
Senes A, Engel DE, DeGrado WF (2004) Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol 14:465–479
Senes A, Gerstein M, Engelman DM (2000) Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol 296:921–936
Senes A, Ubarretxena-Belandia I, Engelman DM (2001) The Cα-H.O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA 98:9056–9061
Stephens SK, Floriano B, Cathcart DP, Bayley SA, Witt VF, Jiménez-Díaz R, Warner PJ, Ruiz-Barba JL (1998) Molecular analysis of the locus responsible for production of plantaricin S, a two-peptide bacteriocin produced by Lactobacillus plantarum LPCO10. Appl Environ Microbiol 64:1871–1877
Uteng M, Hauge HH, Markwick PRL, Fimland G, Mantzilas D, Nissen-Meyer J, Muhle-Goll C (2003) Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and a sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry 42:11417–11426
van Belkum MJ, Hayema BJ, Jeeninga RE, Kok J, Venema G (1991) Organization and nucleotide sequence of two lactococcal bacteriocin operons. Appl Environ Microbiol 57:492–498
Wang Y, Henz ME, Fregeau Gallagher NL, Chai S, Gibbs AC, Yan LZY, Stiles ME, Wishart DS, Vederas JC (1999) Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry 38:15438–15447
Zendo T, Koga S, Shigeri Y, Nakayama J, Sonomoto K (2006) Lactococcin Q, a novel two-peptide bacteriocin produced by Lactococcus lactis QU 4. Appl Environ Microbiol 72:3383–3389
Acknowledgments
This work has been supported by the Norwegian Research Council.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Nissen-Meyer, J., Oppegård, C., Rogne, P. et al. Structure and Mode-of-Action of the Two-Peptide (Class-IIb) Bacteriocins. Probiotics & Antimicro. Prot. 2, 52–60 (2010). https://doi.org/10.1007/s12602-009-9021-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-009-9021-z