Abstract
Background
Prediction modelling can greatly assist the health-care professionals in the management of diseases, thus sparking interest in neonatal sepsis diagnosis. The main objective of the study was to provide a complete picture of performance of prediction models for early detection of neonatal sepsis.
Methods
PubMed, Scopus, CINAHL databases were searched and articles which used various prediction modelling measures for the early detection of neonatal sepsis were comprehended. Data extraction was carried out based on Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies checklist. Extricate data consisted of objective, study design, patient characteristics, type of statistical model, predictors, outcome, sample size and location. Prediction model Risk of Bias Assessment Tool was applied to gauge the risk of bias of the articles.
Results
An aggregate of ten studies were included in the review among which eight studies had applied logistic regression to build a prediction model, while the remaining two had applied artificial intelligence. Potential predictors like neonatal fever, birth weight, foetal morbidity and gender, cervicovaginitis and maternal age were identified for the early detection of neonatal sepsis. Moreover, birth weight, endotracheal intubation, thyroid hypofunction and umbilical venous catheter were promising factors for predicting late-onset sepsis; while gestational age, intrapartum temperature and antibiotics treatment were utilised as budding prognosticators for early-onset sepsis detection.
Conclusion
Prediction modelling approaches were able to recognise promising maternal, neonatal and laboratory predictors in the rapid detection of early and late neonatal sepsis and thus, can be considered as a novel way for clinician decision-making towards the disease diagnosis if not used alone, in the years to come.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neonatal sepsis is the third most prominent cause of mortality among neonates after intrapartum complications and prematurity. Nearly, 13% of neonatal mortality is bequeathed by it, of which 42% of the death occurs in the first week itself [1]. The primary challenge in neonatal sepsis is its evasive signs and symptoms which makes the diagnosis and prognosis burdensome. The only unrivalled quick fix is blood culture confirmation which takes virtually two days to generate result [2]. The necessity of a distinguished biomarker is fundamental for meticulous and for the nick of time diagnosis [3]. Inadequate immunity in neonates makes it soldier to overcome mortality [4]. Therefore, there is a need to glance at novel approaches to embark upon the situation.
Prediction modelling is a statistical measure of employing established results to generate, design and validate a model that can be applied for anticipating expected outcomes [5]. It can greatly assist clinician as well as the health-care professionals on efficient management of any disease [6]. Severity score generated by these prognostic prediction models aids in scrutinising the profoundness of the disease in due time. Furthermore, it recuperates any disease management by risk and patient stratification [7].
Prediction modelling has reinforced in the pronouncement of several potential biomarkers in the diagnosis of neonatal sepsis which essentially comprehend C-Reactive Protein (CRP), Interleukin-27, neutrophil CD64, etc. [8,9,10]. It also inculcates myriad maternal and neonatal risk factors such as intrapartum temperature, gestational age at delivery, duration of premature rupture of membrane (PROM), intrapartum antibiotic treatments, mode of delivery, birth weight, etc. in the conjecture of the disease [11,12,13]. Similarly, clinical and laboratory biomarkers such as maternal white blood cells, absolute neutrophil count was also used for the diagnosis of the disease [13]. Incorporation of laboratory diagnostic markers such as WBC along with standard biomarker such as CRP in the prediction model has considerably reduced the applicability of antibiotics in early-onset neonatal sepsis [14]. Sepsis risk calculator, scoring system generation were some of the desired results of prediction modelling implementation [15, 16]. Similarly, application of sepsis risk calculator through prediction model development also resulted in decrement in the usage of antibiotic therapy [15].
Hence, a systematic review is required to assess this modernistic approach which will give a fresh insight in the prognosis of having neonatal sepsis. The principal purpose of the study is to provide an overall depiction of the entire prediction modelling measures projecting the early detection of neonatal sepsis. As per our proficiency, this would be the aboriginal systematic review to comprehend this facet. The current systematic review will benefit the young researchers and investigators to have an eye at the multifarious pathway to undertake their research in neonatal sepsis meanwhile making a provision for the clinician for improvised supervision on the disease.
Methods
Information sources
Articles were searched from distinct databases such as PubMed, Scopus, and CINAHL using the following keywords “Prediction model” “neonatal sepsis” “neonatal sepses”. Earliest ten-year papers were searched till September 2020 for the study purpose in all the databases.
Eligibility criteria
Papers were comprehended if they fit the following inclusion criteria: (1) Operational definition of neonatal sepsis: Neonates having positive blood culture before 72 h of life were considered as early neonatal sepsis, whereas neonates having positive blood culture report after 72 h of life were termed as late neonatal sepsis. Meanwhile, neonates having positive blood culture report before 30 days of life were considered as neonatal sepsis patient [17]; (2) Case–control, cohort—prospective/retrospective studies predicting the prognosis of having neonatal sepsis (early/late onset) which is to be culture positive; (3) Prediction model developed through various statistical procedure like machine learning/logistic regression/artificial intelligence for early detection of neonatal sepsis; (4) The prediction developmental model should be either internally or externally validated, and (5) Articles published in English peer reviewed journal.
However, randomised clinical trial, review/systematic review/meta-analysis related to early/late neonatal sepsis were excluded from the study. Implementation of the prediction models pertaining to neonatal sepsis or prognosis of the disease was also ruled out from the study.
Study selection
The study screening was conducted in accordance with the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) guidelines [18]. The first reviewer [P.S] evaluated all the search results. Further title, abstract and subsequently full text screening were cross checked by the rest two reviewers (EAR, VK). Title, abstract and full text screening was consummated on the ground of study design/disease/outcome/intervention/irrelevant. Any difference in the opinion regarding final included articles was resolved through discussions.
Data extraction and quality assessment
Data extraction was carried out by all the reviewers based on Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies (CHARMS) checklist [19, 20]. Extricate data consisted of objective, study design, patient characteristics, type of statistical model, predictors in the model, outcome measure, sample size and location. Significant predictors were identified by relative risk or odds ratio. Outcome measures were assessed by area under the curve, sensitivity, specificity, positive predictive and negative predictive value. Prediction model risk of bias assessment tool (PROBAST) was applied to gauge the risk of bias of the included articles and were classified into high, low, and unclear risk of bias. PROBAST risk of bias consists of questions from four domains which include participants, predictors, outcome and analysis. A subtotal of 20 questions were present for the judgement of the paper. These questions were answered as yes/no/probably yes/probably no/ unclear, where “yes” implies the absence of bias, while “no” signifies the presence of bias. An article is denoted as “low risk of bias”, if all the desirable answers to the 20 questions were yes or else it is designated as high risk. An unclear answer to any of the 20 questions represents the article to be of unclear risk of bias [21].
Data synthesis and analysis
Systematic analysis of all the included studies was conducted to generate all the desired data. Statistical measure employed for building the model was determined to figure out the type of it. Predictors were viewed to identify its potential in the model. Performance of the prediction model were determined by noting the major indices like Area Under the curve (AUC), Specificity, Sensitivity, Negative Predictive Value (NPV) and Positive Predictive Value (PPV). External and internal validation of the models was also scrutinised to determine the efficiency of the model.
Results
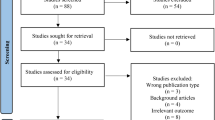
The study screening was conducted in accordance with the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) guidelines. A total of 3135 articles were retrieved from three databases, i.e. PubMed, CINAHL and Scopus. An aggregate of ten studies were incorporated for systematic review after scrutinising on the basis of eligibility criteria. The screening process has been detailed in Fig. 1.
Study characteristics
Majority (five studies) of the included articles were performed in United State of America [24, 26, 29, 30], while the remaining each were from Israel, China, Mexico, United Kingdom and Canada [22, 23, 27, 28, 31]. The highest sample size of all the included studies was 4794, while the minimal is 36, which are illustrated in Fig. 2. Most of the studies were single centre (six studies), while 30% (three studies) were of multicentre origin. Greater number of studies (seven studies) had applied logistic regression to build a prediction model, while two of the studies had applied artificial intelligence. Autoregressive hidden Markov model (AR-HMM) was applied by a single study for prediction model building [28]. Partly, the studies (five studies) had applied split validation (70:30 ratio) concepts for the derivation and validation of the model, while one of the studies had applied external validation [21]; while the other four studies had utilised bootstrap or concordance statistics methodology for internal validation.
Neonatal sepsis detection was the primary outcome of considerable number of studies (five studies) [27,28,29,30,31]. Early-onset and late-onset sepsis detection were encountered as the main outcome in three and two different studies, respectively [22,23,24,25,26]. Sheer single study had stratification of the early-onset neonatal sepsis patients based on the degree of illness as the defined outcome [25]. The length of the age of all the participants during outcome assessment varied from 0 to 30 days of life. The major characteristics of the included studies are summarised in Tables 1, 2 and 3. Predictors adopted in the comprehended studies for constructing the model were classified into four domains such as neonatal factors, maternal factors, clinical, laboratory parameters and miscellaneous. Table 4 represents the predictors notified in each study for the final model prediction.
Risk of bias assessment
Prediction model risk of bias assessment tool (PROBAST) was applied to appraise the risk of bias of the included articles. Six articles were of low risk of bias while two were of high due to their minimal sample size. Remaining two articles were of unclear risk of bias as “analysis domain” of the PROBAST table was unable to give a clear conclusion. Since, one of the domains was unclear, we labelled the paper as the same as per the PROBAST guidelines [21]. The detailed assessment of risk of bias for the included articles using PROBAST tool, based on individual domain is provided in the supplementary table 1.
Predictors’ assessment
Predictors’ assessment of the included articles was evaluated on the basis of neonatal factors, maternal factors, clinical/laboratory parameters with sign and symptoms and miscellaneous factor for quick detection of early, late or neonatal sepsis as a whole.
Maternal factors like maternal age, cervicovaginitis were the potential predictors in detection of sepsis as whole [27]; whereas, group B streptococcus status, duration of premature rupture of membrane aids in evaluating early-onset sepsis [24].
Sepsis detection can be eased out by neonatal predictors like fever, birth weight, foetal morbidity and gender [27]. Birth weight, endotracheal intubation, thyroid hypofunction, umbilical venous catheter (UVC) was the promising neonatal factors for predicting late-onset sepsis [23]; while gestational age, intrapartum temperature and antibiotics treatment were utilised as budding prognosticator for early-onset sepsis detection [24].
Clinical and laboratory parameters suchlike sick appearance, neutrophil–lymphocyte ratio > 1.5, and C-Reactive Protein > 0.75 were able to detect late-onset sepsis at the earliest [22]. Signs and symptoms such as increase in the number of bradycardia and mini-bradycardia events was a reflection on fast onset of neonatal sepsis [28]. Moreover, platelet count, bradypnea, band cells heart rate (maximum, minimum), SpO2 minimum, blood pressure systolic (minimum, mean), thyroid, acyl carnitine, amino acids and adrenal functions were few of the indicators which also showed significant results in the early detection of neonatal sepsis as whole [27,28,29,30].
Miscellaneous factors like oximeter error did not show any significant result [28]; while, catheter presence showed positive sign in fast detection of neonatal sepsis [25].
Outcome measures
The AUC (derivation model AUC = 0.92, validation model AUC = 0.90) noted for the model developed through clinical and laboratory parameters for the prognosis of having late-onset neonatal sepsis was quite satisfactory [22]. Formation and testing of various nomogram from the derivation and validation set (derivation AUC = 0.855, validation AUC = 0.834) for prediction of late-onset neonatal sepsis was also successful in doing so [23].
Intrapartum maternal risk factors were utilised to form a prediction model for early-onset sepsis detection with AUC of 0.807 and 0.794 from derivation and validation set, respectively [24]. Early onset sepsis identification by neural network model had AUC 92.5, accuracy of 86.74, sensitivity 80.32, and specificity of 90.4 [26]. Quantitative stratification algorithm for early-onset sepsis detection had classified neonates with sepsis risk at birth into three categories, i.e. clinical illness, equivocal and well appearing [25].
Autoregressive hidden Markov model with AUC of 0.74 clearly exhibited that physiological change in events such as bradycardia or mini-bradycardia can detect neonatal sepsis prior to culture report [26]. Maternal and neonatal factor model had predicted the neonatal sepsis disease with an AUC of 97.4, specificity 80%, sensitivity 93.33%, PPV 82.35 and NPV of 92.3 which had surpassed the performance of physician also [27]. Non-invasive model developed for early detection of neonatal sepsis through logistic regression showed better results when compared to that of the invasive model (AUC of invasive model 0.777, AUC of non-invasive model 0.824) [30]. Neonatal sepsis detection by another team of authors also displayed similar results (AUC 0.879) [29]. Newborn screening results like thyroid and adrenal function, acyl carnitine and amino acids also improved the model fit for detection of neonatal sepsis [31].
Validation measures
Half of the included studies had utilized 2:1 ratio, i.e. split method of derivation and validation set for evaluating the accuracy, precision and validity of model [22, 24, 25, 29, 30]; meanwhile, mere one study had used the external validation procedure of conducting the same study in different set of samples at different period of time [23]. 70:15:15 (70 derivation set, 15 validations set, 15 testing set) ratio was applied for validation of model developed by maternal and neonatal risk factor for identification of neonates with sepsis. Back propagation and neural network methods were applied to do so. The performance of the model was calculated on three metrics, i.e. root mean square error (RMSE), regression coefficient and statistical slope and intercept [27]. Similar back propagation method was applied by another study for validation of model for early-onset sepsis detection [26]. Average precision and F score was generated by precision recurve call for validation purpose in model built by autoregressive hidden Markov model [28]. Bootstrap method of internal validation (200 samples) was applied to validate model which predicted neonatal sepsis through the analysts of newborn screening [31].
Discussion
Failure to quickly diagnose neonatal sepsis, primarily due to its indefinite sign and symptom makes the disease more lethal and destructive. Blood culture report being the only chief solution, virtually takes two days to generate result. Therefore, there is a need to look into novel approaches, which may help in fast prediction of neonatal sepsis. This systematic review tried to capture the modernistic prediction modelling methods implied for quick detection of late/early/neonatal sepsis as whole, thus endowing a fresh insight to the approach for disease management. Thus, the main purpose of our study was to perform a systematic review which would give a detailed illustration of prediction modelling measure in the prognosis of having neonatal sepsis. To the best of our knowledge, none of the available studies had focussed on the prediction modelling methods in neonatal sepsis. To fixate our study in prediction modelling in prognosis of having neonatal sepsis, the current review illustrations were based on the foundation of three divisions, i.e. principal findings, predictors assessed, comparison among included articles based on statistical approach implied to build the model.
Principal findings
This systematic review illustrated that few articles were available on prediction modelling for fast detection of early/late/neonatal sepsis as whole. Out of those, most articles included had displayed satisfactory results in terms of prediction of the disease. Maternal, neonatal, clinical and laboratory predictors identified through these models will assist health-care professionals in the rational management of the disease [22,23,24, 27, 29, 30]. Model performance for quick detection of sepsis was almost equivalent to that of physician performance [27]. Stratification of early-onset sepsis into clinically ill, equivocal and well appearing using prediction modelling had helped in modifying the treatment approach for individual categories [25]. Physiological events like increased number of bradycardia or mini-bradycardia were identified as good predictors to detect the disease prior to the culture report [28]. Thus, it can be concluded that prediction modelling measures can be used as an additional approach to clinician decision, if not used alone.
Predictors assessed compared with other studies
Included articles in our study had either utilised clinical/laboratory parameters, neonatal/maternal risk factors, or physiological events of the neonates for building the prediction model [22,23,24,25,26,27,28,29,30,31]. Birth weight characteristics, gestational age features, role of setting, sampling strategy and concomitant interventions were some of the primary predictors assessed for late-onset neonatal sepsis by routine neonatal screening [34]. Analogous results were found in one of the included articles where birth weight, endotracheal intubation, thyroid hypofunction and umbilical venous catheter (UVC) were the promising neonatal factors for predicting late-onset sepsis [23]. Study conducted by Seidel et al. for predicting late-onset sepsis by body surface screening had pooled sensitivity of 41% (95% CI 17–70%) and specificity of 56% (95% CI 34–76%), whereas another included study in our review for predicting late-onset sepsis by clinical and laboratory evaluation had shown sick appearance (OR 5.7, 95% CI 1.1–29.1), CRP > 0.75 (OR 5.4, 95% CI 1.1–26.3)), neutrophil to lymphocyte ratio > 1.5 (OR 6.7, 95% CI 1.2–38.5) as the chief predictors with model AUC of 0.92 [22].
Another systematic review was designed to evaluate the outcome of early neonatal sepsis by sepsis calculator, and had shown reduced antibiotic usage and fewer surges in mortality and readmission to hospital [32, 33]; whereas, one of the included articles in the review had different objective and had shown gestational age, intrapartum temperature and antibiotics treatment as budding prognosticator for early-onset sepsis detection [24]. Yet another included article had classified early-onset sepsis patients into various categories based on the degree of illness. Stratification of those patients had enabled the health-care professionals to render distinct treatment to various categories, thus ensuring proper management [25].
Study conducted by Liang et al. illustrated that prematurity, low birth weight and young age at presentation were some of the major factors which were associated with mortality in neonates with sepsis [35]. Artificial neural network approach was employed by another included article for analysing various maternal and neonatal predictors for early detection of neonatal sepsis and had suggested that maternal age, neonatal fever, apnoea, platelet count was the most valuable predictor of sepsis followed by cervicovaginitis, gender, bradypnea, band cells, catheter presence, birth weight, neutrophil counts and foetal morbidity.
Apart from the previously mentioned predictors, novel diagnostic marker such as neutrophil CD64, platelet to lymphocyte and neutrophil to lymphocyte can also be added to the prediction model for increasing the accuracy of diagnosis of neonatal sepsis as neutrophil CD64 when combined with standard biomarkers like CRP and WBC increases the sensitivity and accuracy of diagnosis [10, 37]. Likewise, inclusion of platelet to lymphocyte and neutrophil to lymphocyte ratio as predictors in the model will increment the diagnosing capacity of early-onset neonatal sepsis in particular [37]. Thus, addition of novel biomarker along with the standard biomarkers in the model will potentiate the predicting ability of the model.
Comparison among included articles based on statistical approach employed to build the model
Among the two of the included articles used for the quick detection of the late-onset neonatal sepsis, Huang et al. had the most appropriate methodology which can be used in other clinical settings for building prediction model [22, 23]. Logistic regression adopted for building the model had several advantages in comparison to the other approaches as it provide probability prediction of each predictor unlike only classifying it into different levels [38]. Models generated by this approach are less likely to be over fit even with the smaller datasets. Unlike other approaches like support vector machines and decision tree, it can easily accommodate larger data set and can refurbish easily to reflect new data set [39]. External validation approach was employed by Huang et al. to measure the accuracy of the model. It ensures reproducibility and transportability of the model in other clinical settings [40, 41]. Being a multicenter origin study, it provides the build model with satisfactory power to detect late-onset sepsis in heterogeneous subjects [42]. Validation cohort AUC of 0.963 of the study also highlighted similar results [23].
Out of all the included articles for fast detection of early-onset neonatal sepsis, Escobar et al.’s study can be considered to have remarkably relevant methodology [24,25,26]. Similar kind of methodology was adopted by Puopolo et al. and Martinez et al. [22, 23]; however, Escobar et al.’s study can be primarily considered as it had stratified early-onset sepsis neonates into clinical illness, equivocal and well appearing. Stratification method adopted in the study will assist the clinician in providing appropriate treatment modalities and strategy for improved patient care in various subgroups [43]. Logistic regression, multicenter origin, and internal validation employed by this study had made it a desirable prediction model; however, external validation was necessary for replicating the methodology in other clinical settings.
Among the five selected articles for early detection of neonatal sepsis, three had used the same statistical measure, i.e. logistic regression [29,30,31], while the remaining one had used autoregressive hidden Markov model approach [28]. Autoregressive Markov model is better than simple Markov model as it identifies a greater number of sequences. It also allows various sequences to be modelled rather than only inserting or deleting it. Unlike simple Markov models, these are quite expensive and require larger seed sequences to be trained thus, is difficult to replicate [44]. Study conducted by Thakur et al. 2019, 2018 was quite similar based on sample size, validation measure and outcome viewed. However, replicating it in other settings is still questionable as external validation was not performed [29, 30]. Bootstrapping method applied by Fell et al. is one of the efficient ways of internal validation when compared to that of cross or split validation as it utilises the entire data set for validating which produces results of low variance [45] still, cannot surpass the advantages of external validation. Thus, external validation being one of the inevitable parts of prediction modelling should be taken into consideration in the future research for the applicability quotient.
Strengths
Articles containing model validation part (internal/external) were particularly included in the review, as models without validation were of low robustness and accuracy and difficult to replicate in other settings [46,47,48].
Limitations
Studies conducted in English language were only considered for the review due to which a number of articles had to be excluded and thus, can be considered as one of the shortcomings. Meanwhile, scarcity of data also affected the outcome of the review. Further research (both interventional and observational studies) is required in this field to generate more evidence.
Thus, the chief findings of this review are that prediction modelling can be considered as a novel way of approach towards the disease, which not only will help the clinician in making rationale and critical decision but will help the young researchers in considering prediction modelling as an important tool for investigation.
Conclusions
Models generated through various statistical approaches were successful in predicting the disease prior to culture report and had attained comparable outcome to that of the physician. Stratification of patient pertaining to severity of illness for varied treatment approach is another key point of the prediction model. Outcome obtained from the various prediction models were successful in generating potential predictors that can benefit the clinician in appropriate management of the disease. Thus, it can be effectively concluded that prediction model developed through proper statistical measures can be recognised as a novel way of approaching towards neonatal sepsis detection in the years to come. However, extensive research is in need to assert the prediction modelling as a sole measure for disease management based on external validation as an aspect of reference and reproducibility.
Data availability
Data sharing is not applicable to this study as no data have been generated or analysed during the present study.
References
Zea-Vera A, Ochoa TJ. Challenges in the diagnosis and management of neonatal sepsis. J Trop Pediatr. 2015;61:1–13.
Iroh Tam PY, Bendel CM. Diagnostics for neonatal sepsis: current approaches and future directions. Pediatr Res. 2017;82:574–83.
Gilfillan M, Bhandari V. Neonatal sepsis biomarkers: where are we now? Res Rep Neonatol. 2019;9:9–20.
Yu JC, Khodadadi H, Malik A, Davidson B, Salles ÉDSL, Bhatia J, et al. Innate immunity of neonates and infants. Front Immunol. 2018;9:1759.
Frankenfield J. What is predictive modelling? Investopedia [Internet] 2020. Available from: www.investopedia.com/terms/p/predictive-modeling.asp. Accessed 27 Dec 2020.
Moons KG, Royston P, Vergouwe Y, Grobbee DE, Altman DG. Prognosis and prognostic research: what, why, and how? BMJ. 2009;338:b375.
Royston P, Moons KG, Altman DG, Vergouwe Y. Prognosis and prognostic research: developing a prognostic model. BMJ. 2009;338:604.
Beltempo M, Viel-Thériault I, Thibeault R, Julien AS, Piedboeuf B. C-reactive protein for late-onset sepsis diagnosis in very low birth weight infants. BMC Pediatr. 2018;18:16.
He Y, Du WX, Jiang HY, Ai Q, Feng J, Liu Z, et al. Multiplex cytokine profiling identifies interleukin-27 as a novel biomarker for neonatal early onset sepsis. Shock. 2017;47:140–7.
Yang AP, Liu J, Yue LH, Wang HQ, Yang WJ, Yang GH. Neutrophil CD64 combined with PCT, CRP and WBC improves the sensitivity for the early diagnosis of neonatal sepsis. Clin Chem Lab Med. 2016;54:345–51.
Van der Ham DP, Van Kuijk S, Opmeer BC, Willekes C, Van Beek JJ, Mulder AL, et al. Can neonatal sepsis be predicted in late preterm premature rupture of membranes? Development of a prediction model. Eur J Obstet Gynecol Reprod Biol. 2014;176:90–5.
Adatara P, Afaya A, Salia SM, Afaya RA, Konlan KD, Agyabeng-Fandoh E, et al. Risk factors associated with neonatal sepsis: a case study at a specialist hospital in Ghana. Sci World J. 2019;2019:9369051.
Popowski T, Goffinet F, Maillard F, Schmitz T, Leroy S, Kayem G. Maternal markers for detecting early-onset neonatal infection and chorioamnionitis in cases of premature rupture of membranes at or after 34 weeks of gestation: a two-center prospective study. BMC Pregnancy Childbirth. 2011;11:26.
Stocker M, Daunhawer I, van Herk W, El Helou S, Dutta S, Schuerman F, et al. Machine learning used to compare the diagnostic accuracy of risk factors, clinical signs and biomarkers and to develop a new prediction model for neonatal early-onset sepsis. Pediatr Infect Dis J. 2021. https://doi.org/10.1097/INF.0000000000003344.
Weber EJ, Sanchez LC, Giguère S. Re-evaluation of the sepsis score in equine neonates. Equine Vet J. 2015;47:275–8.
Achten NB, Klingenberg C, Benitz WE, Stocker M, Schlapbach LJ, Giannoni E, et al. Association of use of the neonatal early-onset sepsis calculator with reduction in antibiotic therapy and safety: a systematic review and meta-analysis. JAMA Pediatr. 2019;173:1032–40.
Singh M, Alsaleem M, Gray CP. Neonatal Sepsis. [Updated 2021 Oct 10]. In: StatPearls [Internet] 2021. Available from: www.ncbi.nlm.nih.gov/books/NBK531478/. Accessed 10 Oct 2020.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Moons KG, de Groot JA, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11:e1001744.
Wee L, van Kuijk SMJ, Dankers F, Traverso A, Welch M, Dekker A. Reporting standards and critical appraisal of prediction models. In: Kubben P, Dumontier M, Dekker A, editors. Fundamentals of clinical data science. Cham (CH): Springer; 2019. p. 135–50.
Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019;170:51–8.
Goldberg O, Amitai N, Chodick G, Bromiker R, Scheuerman O, Ben-Zvi H, et al. Can we improve early identification of neonatal late-onset sepsis? A validated prediction model. J Perinatol. 2020;40:1315–22.
Huang Y, Yu X, Li W, Li Y, Yang J, Hu Z, et al. Development and validation of a nomogram for predicting late-onset sepsis in preterm infants on the basis of thyroid function and other risk factors: mixed retrospective and prospective cohort study. J Adv Res. 2020;24:43–51.
Puopolo KM, Draper D, Wi S, Newman TB, Zupancic J, Lieberman E, et al. Estimating the probability of neonatal early-onset infection on the basis of maternal risk factors. Paediatrics. 2011;128:1155–63.
Escobar GJ, Puopolo KM, Wi S, Turk BJ, Kuzniewicz MW, Walsh EM, et al. Stratification of risk of early-onset sepsis in newborns ≥ 34 weeks’ gestation. Pediatrics. 2014;133:30–6.
López-Martínez F, Núñez-Valdez ER, Lorduy Gomez J, García-Díaz V. A neural network approach to predict early neonatal sepsis. Comput Electr Eng. 2019;76:379–88.
Helguera-Repetto AC, Soto-Ramírez MD, Villavicencio-Carrisoza O, Yong-Mendoza S, Yong-Mendoza A, León-Juárez M, et al. Neonatal sepsis diagnosis decision-making based on artificial neural networks. Front Pediatr. 2020;8:525.
Stanculescu I, Williams CK, Freer Y. Autoregressive hidden Markov models for the early detection of neonatal sepsis. IEEE J Biomed Health Inform. 2014;18:1560–70.
Thakur J, Pahuja S, Pahuja R. Non-invasive prediction model for developing countries to predict SEPSIS in neonates. Biomed Eng Appl Basis Commun. 2019;31:1950001.
Thakur J, Pahuja SK, Pahuja R. Neonatal sepsis prediction model for resource-poor developing countries. 2nd International Conference on Electronics, Materials Engineering & Nano-Technology (IEMENTech) 2018:1–5.
Fell DB, Hawken S, Wong CA, Wilson LA, Murphy MSQ, Chakraborty P, et al. Using newborn screening analytes to identify cases of neonatal sepsis. Sci Rep. 2017;7:18020.
Achten NB, Klingenberg C, Benitz WE, Stocker M, Schlapbach LJ, Giannoni E, et al. Association of use of the neonatal early-onset sepsis calculator with reduction in antibiotic therapy and safety: a systematic review and meta-analysis. JAMA Paediatr. 2019;173:1032–40.
Deshmukh M, Mehta S, Patole S. Sepsis calculator for neonatal early onset sepsis—a systematic review and meta-analysis. J MaternFetal Neonatal Med. 2021;34:1832–40.
Harder T, Seidel J, Eckmanns T, Weiss B, Haller S. Predicting late-onset sepsis by routine neonatal screening for colonisation by gram-negative bacteria in neonates at intensive care units: a protocol for a systematic review. BMJ Open. 2017;7:e014986.
Liang LD, Kotadia N, English L, Kissoon N, Ansermino JM, Kabakyenga J, et al. Predictors of mortality in neonates and infants hospitalized with sepsis or serious infections in developing countries: a systematic review. Front Pediatr. 2018;6:277.
Seidel J, Haller S, Eckmanns T, Harder T. Routine screening for colonization by Gram-negative bacteria in neonates at intensive care units for the prediction of sepsis: systematic review and meta-analysis. J Hosp Infect. 2018;99:367–80.
Zhang S, Luan X, Zhang W, Jin Z. Platelet-to-lymphocyte and neutrophil-to-lymphocyte ratio as predictive biomarkers for early-onset neonatal sepsis. J Coll Physicians Surg Pak. 2021;30:821–4.
Collins GS, Ogundimu EO, Altman DG. Sample size considerations for the external validation of a multivariable prognostic model: a resampling study. Stat Med. 2016;35:214–26.
Advantages and Disadvantages of Logistic Regression [Internet]. OpenGenus IQ: Computing Expertise & Legacy. 2021. Available from: https://iq.opengenus.org/advantages-and-disadvantages-of-logistic-regression/. Accessed 25 Nov 2021.
Logistic Regression Pros & Cons - HolyPython.com [Internet]. HolyPython.com. 2021. Available from: https://holypython.com/log-reg/logistic-regression-pros-cons. Accessed 25 Nov 2021.
König IR, Malley JD, Weimar C, Diener HC, Ziegler A. Practical experiences on the necessity of external validation. Stat Med. 2007;26:5499–511.
Chung KC, Song JW. A guide to organizing a multicenter clinical trial. Plast Reconstr Surg. 2010;126:515–23.
Casagranda I, Vendramin C, Callegari T, Vidali M, Calabresi A, Ferrandu G, et al. Usefulness of suPAR in the risk stratification of patients with sepsis admitted to the emergency department. Intern Emerg Med. 2015;10:725–30.
Strengths and weaknesses of hidden Markov models [Internet]. Compbio.soe.ucsc.edu. 2021. Available from: https://compbio.soe.ucsc.edu/html_format_papers/tr-94-24/node11.html. Accessed 25 Nov 2021.
Bleeker SE, Moll HA, Steyerberg EW, Donders AR, Derksen-Lubsen G, Grobbee DE, et al. External validation is necessary in prediction research: a clinical example. J Clin Epidemiol. 2003;56:826–32.
Mayer D, Butler D. Statistical validation. Ecol Model. 1993;68:21–32.
Collins GS, de Groot JA, Dutton S, Omar O, Shanyinde M, Tajar A, et al. External validation of multivariable prediction models: a systematic review of methodological conduct and reporting. BMC Med Res Methodol. 2014;14:40.
Moons KG, Kengne AP, Woodward M, Royston P, Vergouwe Y, Altman DG, et al. Risk prediction models: I, Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 2012;98:683–90.
Acknowledgements
We want to thank Mr. Sawan Mukund for his contribution in reviewing and making figures of the manuscript. This study was supported by no funding; however, the fellowship for the research participant was supported by Indian Council of Medical Research (ICMR).
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal. No funding was supported for this study.
Author information
Authors and Affiliations
Contributions
PS and EAR conceptualised and designed the study, extracted and analysed the data, drafted the initial manuscript and reviewed and revised the manuscript. VK designed and supervised the data extraction procedure, chooses and scrutinised risk of bias assessment tool, supervised data analysis and reviewed and revised the manuscript. LESL, KP, MR had supervised data extraction and analysis procedure and had critically reviewed the manuscript for intellectual content. All authors approved the final manuscript as submitted and agreed to be accountable for all the aspect of the work.
Corresponding author
Ethics declarations
Ethical approval
Not required for a systematic review.
Conflict of interest
No financial or non-financial benefits have been received or will be received from any party related directly or indirectly to the subject of this article. None of the authors were involved in the journal's review of, or decisions related to, this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sahu, P., Raj Stanly, E.A., Simon Lewis, L.E. et al. Prediction modelling in the early detection of neonatal sepsis. World J Pediatr 18, 160–175 (2022). https://doi.org/10.1007/s12519-021-00505-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12519-021-00505-1