Abstract
Chemokines and their receptors are implicated in formation of colorectal cancer metastases. Especially CXCR4 is an important factor, determining migration, invasiveness, metastasis and proliferation of colorectal cancer cells. Object of this study was to determine expression of CXCR4 in tumor tissue of colorectal cancer patients and associate CXCR4 expression levels to clinicopathological parameters. Levels of CXCR4 expression of a random cohort of patients, who underwent primary curative resection of a colorectal carcinoma, were retrospectively determined by quantitative real-time RT-PCR and semi-quantitative analyses of immunohistochemical stained paraffin sections. Expression levels were associated to clinicopathological parameters. Using RT-PCR we found that a high expression of CXCR4 in the primary tumor was an independent prognostic factor for a poor disease free survival (p = 0.03, HR: 2.0, CI = 1.1–3.7). Immunohistochemical staining showed that nuclear distribution of CXCR4 in the tumor cells was inversely associated with disease free and overall survival (p = 0.04, HR: 2.6, CI = 1.0–6.2), while expression in the cytoplasm was not associated with prognosis. In conclusion, our study showed that a high expression of nuclear localized CXCR4 in tumor cells is an independent predictor for poor survival for colorectal cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More than 100 years after postulation of the “seed and soil” theory, the precise mechanisms determining the directional migration and invasion of diminished cancer cells into specific organs remain to be established [1, 2]. Recent studies increasingly show that chemokines and their receptors are an important factor in this process of organ selective metastasis [3].
Chemokines are small signaling cytokines that act as chemoattractants through interaction with G-protein-coupled, seven transmembrane domain receptors [4, 5]. They are the major regulators of cell trafficking and adhesion. Specific chemokines are produced and released by target organs that attract tumor cells with specific corresponding receptors, resulting in site/organ specific cancer cell migration and formation of metastasis. This migration signaling mechanism is supported by studies in cancer models, demonstrating that malignant cells can target specific organs or tissues by selected chemokine receptor-ligand interaction [6–10]. Accordingly, neutralization of CXCL12-CXCR4 interaction leads to a marked inhibition of metastasis in tumor animal models [6, 11, 12].
Muller et al. were the first to implicate a key role for CXCR4-CXCL12 in the organ specific metastasis of breast cancer [6]. Thereafter, numerous authors have reported on the involvement of CXCR4-CXCL12 in promoting the metastatic homing of different types of tumor cells, including colorectal cancer [10, 13–16]. CXCR4 is expressed in intestinal cells and over-expressed in colorectal tumor cells [16–18]. It is activated upon binding with its ligand CXCL12 also known as stromal cell-derived factor (SDF-1), triggering cell adhesion, directional migration and proliferation of tumor cells [6]. CXCL12 is normally produced by stromal cells of lymph nodes, lung, liver and bone marrow. These are the most frequent sites for colorectal cancer metastases [19].
At the moment only the TNM classification is used to stage patients with colorectal cancer. New prognostic biomarkers are required to improve staging of colorectal cancer patients and thereby resulting in better selection of patients that might benefit from (adjuvant) therapy. Many studies have demonstrated an important association between CXCR4 expression and clinical prognosis of patients with various types of cancer [3, 13, 14, 20–23]. In our study, we retrospectively determined the level of expression and cellular distribution of CXCR4 in association with clinical, pathological and prognostic parameters in tumor tissue of a random selected cohort of colorectal cancer patients, using RT-PCR and immunohistochemical techniques. This study focuses whether CXCR4 might function as a biomarker to improve the current staging of colorectal cancer patients.
Material and Methods
Tumor Specimens for RT-PCR
RNA from snap-frozen tumor samples, containing at least 60% tumor cells as determined by a pathologist (HM), of 70 curatively operated colorectal cancer patients was isolated using RNeasy columns (Qiagen, USA). All patient materials were obtained with approval of local medical ethic committee. Patients were operated between 1990 and 2001, at the time of censoring 41 (59%) had died of whom 22 (54%) died from their disease, and 29 patients were still alive; four of them were alive with recurrence of the tumor. Mean follow up was 99 months (range 50–172 months). Patients with stage I/II (n = 47) and stage III (n = 23) colorectal cancer (as defined by the American Joint committee on Cancer and Union Internationale Contre le Cancer-criteria) were selected for this study.
RT-PCR of CXCR4 in a Patient Cohort
PCR primers for the detection of CXCR4 and the house-keeping genes (heterogeneous nuclear ribonucleoprotein M (HNRPM) and TATA box binding protein (TBP) were designed in PRIMER Express (Applied Biosystems, USA) and span at least one exon-exon boundary. The primers used were: HNRPM, 5’-GAGGCCATGCTCCTGGG-3’, 5’-TTTAGCATCTTCCATGTGAAATCG-3’, TBP, 5’-CACGAACCACGGCACTGAT-3’, 5’-TTTTCTTGCTGCCAGTCTGGAC-3’ and CXCR4 5’-TTCTACCCCAATGACTTGTG-3’-5’-ATGTAGTAAGGCAGCCAACA-3’. RT-PCR reactions were performed on an ABI Prism 7900ht (Applied Biosystems) using the SybrGreen RT-PCR core-kit (Eurogentec, Belgium). Cycle conditions were 10 min at 95°C followed by 40 cycles of 10 s at 95°C and 1 min at 60°C. Cycle threshold extraction was performed using the SDS software (version 2.2.2, Applied Biosystems). For all PCR reactions, a standard curve was generated using a five-step, five-fold dilution of pooled cDNA from the HCT81 colorectal cancer cell line. Relative concentrations of mRNA for each gene were calculated from the standard curve. After RT-PCR, dissociation curves were made to check the quality of the reaction. Reactions with more than one peak in the dissociation curve were discarded. For normalization, the expression values for each gene were divided by the normalization factor of the gene (the average of the two house keeping genes).
Immunohistochemistry of CXCR4 in a Patient Cohort
A tissue microarray (TMA) was constructed from formalin-fixed, paraffin-embedded tissues from 58 curatively operated colorectal cancer patients as described previously [24]. Standard three-step, indirect immunohistochemistry was performed on 4-μm tissue sections transferred to glass slides using a tape-transfer system (Instrumedics, USA), including citrate antigen retrieval and blockage of endogenous peroxidase. Sections were overnight incubated with the primary antibody CXCR4 (Mouse-anti-Human CXCR4 IgG2B, clone MAB172, R&D Systems, USA). Secondary reagent used was biotinylated rabbit anti-mouse IgG antibodies (DAKO Cytomation, Denmark) and biotinylated-peroxidase streptavidin complex (SABC; DAKO Cytomation, Denmark). Microscopic analysis was assessed by two independent observers (F.M.S. and C.J.K.) in a double-blinded manner. Three different punches per patient were scored. Cytoplasmic and nuclear intensity of CXCR4 staining were separately scored in two categories: low (0) or strong (1). Since the envelope of all nuclei of all tumors was stained, nuclear intensity was determined on the degree of staining of the nucleoplasm. Where discrepancies arose between the staining of scores from the same tumor, an average of the scores was taken, with confirmation by two observers using a double-headed microscope with a consensus decision taken in all cases. Tissue stromal cells, normal epithelium or lymph follicles served as positive internal controls to ascertain the quality of the staining. To distinct microsatellite instable (MSI) from microsatellite stable (MSS) tumors, the TMA was stained for mismatch repair proteins MLH1 and PMS2, as previously described [25]. MLH1 and PMS2 are deficient in sporadic MSI tumors. Therefore, the expression of these proteins was used to differentiate MSI and MSS rectal cancers. Tissue stromal cells, normal epithelium or lymph follicles served as positive internal controls when analyzing MLH1, PMS2 expression. The expression of MLH1 and PMS2 was scored positive if tumor cells showed expression, and negative if tumor cells showed no expression of either MLH1 or PMS2, provided that tissue stromal cells did show expression, indicating MSS and MSI tumors, respectively [26].
Statistical Analysis
All analyses were performed with SPSS statistical software (version 12.0 for Windows, SPSS Inc, Chicago, USA). Mann-Whitney U test (M-W) was used to compare variables. Kaplan-Meier analyses were performed to analyze patient survival. The entry date for the survival analyses was the time of surgery of the primary tumor. Events for disease free survival and overall survival were defined as follows: from time of surgery to time of disease relapse or death by any cause (for disease free survival) and time of death by any cause (for overall survival), respectively. Patients were first separately analyzed in univariate analysis in addition, variables with a P value of <0.10 in the univariate analyses were subjected to a multivariate analysis. Cox’ regression analyses were used to calculate hazard ratios (HR) with 95% confidence intervals (CI).
Results
Low Levels of CXCR4 RNA Expression Predict Good Prognosis
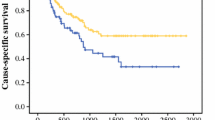
The RNA level of CXCR4 was determined in primary tumor tissue of a cohort of 70 colorectal cancer patients using quantitative RT-PCR and linked to clinical follow-up data. The impact of high versus low expression of CXCR4 was assessed using the 50th percentile cut-off point as previously defined [10, 14]. The characteristics of the cohort colorectal cancer patients, included in this study are summarized in Table 1. To evaluate whether CXCR4 and clinicopathological features were associated, the level of CXCR4 was correlated to each feature. CXCR4 expression was not associated with any of the clinicopathological variables (Table 1). Univariate cox regression analyses were performed to identify prognostic factors for disease free survival and overall survival (Table 1). Advanced patient age (p = 0.006; p = 0.005), TNM stage (p < 0.001; p < 0.001), and high CXCR4 expression (p = 0.006; p = 0.01) proved to be significant predictors for poor disease free and overall survival respectively, using univariate analyses (Table 1). The Kaplan-Meier curve for disease free survival plotting high versus low expression of CXCR4 is shown in Fig. 1. High expression of CXCR4 retained its strength as independent predictor of decreased prognosis in disease free survival (HR: 2.0, p = 0.03; Table 1). Also, TNM stage (HR: 2.9, p = 0.001; HR: 3.1, p = 0.001) retained its strength as independent predictors for disease free and overall survival, while patient age (HR: 2.0, p < 0.05) was found to be an independent predictor only for overall survival. Our RT-PCR results showed that high expression of CXCR4 is independently associated with poor disease free survival for colorectal cancer patients.
Correlation between disease free survival and expression of CXCR4 assessed by RT-PCR in a cohort of colorectal cancer patients.Kaplan Meier survival curve is displayed. Patients with low expression of CXCR4 had a significant (p = 0.006) increased disease free survival compared to patients with high expression of CXCR4
Nuclear Localization of CXCR4 Determines Prognosis for Colorectal Cancer Patients
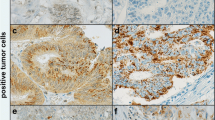
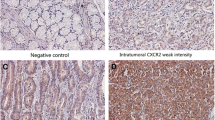
Using immunohistochemistry a TMA of 58 colorectal tumors was stained for CXCR4. We observed immunoreactivity for CXCR4 in the cytoplasm, cell membrane and nucleus of normal and tumor intestinal epithelial cells (Fig. 2). For prognostic purpose only CXCR4 expression in the cancer epithelium was scored. Cytoplasmic staining and nuclear staining were semi-quantitative analyzed, according to previous publications [20]. For cytoplasmic CXCR4 staining 22 (38%) tumors were classified as weak and 36 as strong (62%). For nuclear CXCR4 staining 15 tumors were classified as low (26%) and 43 were strong (74%). No correlation was found between nuclear and cytoplasmic expression of CXCR4. Also no correlation was found between level of CXCR4 mRNA and either nuclear or cytoplasmic expression of CXCR4 as determined by immunohistochemical techniques. Association of cytoplasmic CXCR4 expression to clinicopathological and survival parameters did not reveal any significant correlation. In contrast to cytoplasmic localized CXCR4, nuclear localized CXCR4 was found to be a significant predictor for survival. Using univariate cox regression analyses, we showed that strong expression of CXCR4 was significantly (p = 0.03) associated with decreased overall survival compared to patients with weak nuclear expression of CXCR4. Patient characteristics and several markers that have an effect on disease free survival and overall survival in colorectal cancer showed no significant association with level of CXCR4 (Table 2). In addition, patient age (p = 0.008, p = 0.006) and TNM stage (p = 0.002, p = 0.002) were found to be significant predictors for disease free survival and overall survival respectively (Table 2). Using cox multivariate analysis, strong expression of CXCR4 (HR: 2.6, p = 0.04; HR: 3.7, p = 0.02) retained its strength as independent predictor for both poor disease free survival and overall survival, together with TNM stage (HR: 2.9, p = 0.003; HR: 3.3, p = 0.002) and median age (HR: 2.5, p = 0.01; HR: 2.8, p = 0.008; Table 2). Semi-quantitative analysis of immunohistochemical staining associated to survival showed that strong nuclear localization was associated with poor prognosis for colorectal cancer patients.
Examples of CXCR4 immunohistochemical staining of human colorectal tumors. a displays an example of weak cytoplasmic staining in combination with strong staining of the nucleoplasm. b displays an example of intermediate cytoplasmic staining in combination with weak nuclear staining for CXCR4. c displays an example of strong cytoplasmic intensity of CXCR4 while of the nucleus only the envelope is stained and the nucleoplasm is not stained; original magnification ×400
Discussion
The expression of CXCR4 has been detected in a large number of different types of cancers, together with its use as prognostic biomarker [3, 27]. In the present study we evaluated the expression of CXCR4 in colorectal cancer by quantitative RT-PCR and immunohistochemical staining. Strong expression of nuclear localized CXCR4 and high RNA levels of CXCR4 were both independent significant predictors for poor overall and disease free survival. Our results were consistent with others’ recent RT-PCR data [10, 15].
We found no correlation between expression of CXCR4 mRNA (RT-PCR) and nuclear CXCR4 expression (immunohistochemistry). This might be explained that level of CXCR4 mRNA does not distinct between levels of membrane expressed CXCR4 protein and nuclear expressed CXCR4. Also does RNA isolated from tumor samples, includes RNA from cells other than tumor cells, for instance tumor infiltrated T cells. Tumor infiltrated T cells also express CXCR4 [28, 29] and presence is positively associated with prognosis of colorectal cancer patients [20–23]. As a result tumor infiltrated T cells might disturb prognostic evaluation of CXCR4 mRNA expression isolated from tumor tissues by quantitative RT-PCR. Therefore we additionally used immunohistochemical techniques to semi-quantitatively assess expression of CXCR4 in tumor cells only. Although RT-PCR is a better technique to quantify level of expression, the use of immunohistochemical techniques for clinical and prognostic purposes is preferred above RT-PCR, since the intratumoral and intracellular distribution of CXCR4 can be determined which is not possible using RT-PCR. For prognostic purposes we showed that only nuclear localization of CXCR4 was independently predictive for prognosis of colorectal cancer patients in contrast to expression in the cytoplasm. Using immunohistochemical staining to semi-quantitatively score nuclear and cytoplasmic expression of CXCR4 and associating results to survival parameters, has been done in various types of tumors amongst others in a large panel of breast carcinomata [20–23]. To our knowledge, only two studies determined the association between colorectal cancer and prognosis, using immunohistochemical techniques [13, 15]. These studies only detected cytoplasmic and sometimes membrane staining, while no nuclear staining was separately investigated in both studies. We observed expression of CXCR4 both in the cytoplasm and nucleus of colorectal cancer tissue and though rarely, membrane expression. Our study is the first that was able to distinguish nuclear from cytoplasmic CXCR4 expression in colorectal cancer. A possible explanation for this fact might be that we used a different antibody compared with previous studies.
Shim et al. showed in cultured cells that CXCL12 ligand binding to CXCR4 induced translocation of CXCR4 to the cytoplasm and to the nucleus of cells [30]. The translocation of CXCR4 to the nucleus might be involved in biological processes and function as a transcription factor as has been described for other receptors, for instance the epidermal growth factor receptor (EGFR) [30, 31]. Recently for lung tumors it has been shown that CXCL12 activates CXCR4 receptor and ERK pathway, which in turn induces IKKa/b phosphorylation, p65 Ser536 phosphorylation, and NF-kB activation, which leads to b1 and b3 integrins expression and increases the migration of human lung cancer cells [32]. Since our data imply that especially nuclear staining predicts prognosis, additional research should provide insight in the nuclear function of CXCR4 in colorectal cancer. Moreover if CXCR4 is a transcription or has another specific function in the nucleus it is important to learn which genes are activated or inhibited by CXCR4 in colorectal cancer cells.
Currently, only the TNM staging is used to stage patients with colorectal cancer. Adjuvant treatment is based on this staging. Combining TNM staging with selected biomarkers might better define patients who are at risk for metastases or recurrences and might define patients who would benefit from adjuvant treatment. In conclusion, our data showed that especially nuclear localized CXCR4 determines prognosis for colorectal patients. The use of CXCR4 might improve the current staging of colorectal cancer patients.
References
Paget S (1889) The distribution of secondary growths in cancer of the breast. Lancet 1:571–573
Fuchs E (1882) Das Sarkom des Uvealtractus. Graefe’s Archiv für Ophthalmologie XII:233
Ruffini PA, Morandi P, Cabioglu N et al (2007) Manipulating the chemokine-chemokine receptor network to treat cancer. Cancer 109:2392–2404
Zlotnik A, Yoshie O, Nomiyama H (2006) The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol 7:243
Bacon K, Baggiolini M, Broxmeyer H et al (2002) Chemokine/chemokine receptor nomenclature. J Interferon Cytokine Res 22:1067–1068
Muller A, Homey B, Soto H et al (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410:50–56
Zeelenberg IS, Ruuls-Van Stalle L, Roos E (2003) The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res 63:3833–3839
Koshiba T, Hosotani R, Miyamoto Y et al (2000) Expression of stromal cell-derived factor 1 and CXCR4 ligand receptor system in pancreatic cancer: a possible role for tumor progression. Clin Cancer Res 6:3530–3535
Taichman RS, Cooper C, Keller ET et al (2002) Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res 62:1832–1837
Kim J, Mori T, Chen SL et al (2006) Chemokine receptor CXCR4 expression in patients with melanoma and colorectal cancer liver metastases and the association with disease outcome. Ann Surg 244:113–120
Lapteva N, Yang AG, Sanders DE et al (2005) CXCR4 knockdown by small interfering RNA abrogates breast tumor growth in vivo. Cancer Gene Ther 12:84–89
Liang Z, Yoon Y, Votaw J et al (2005) Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res 65:967–971
Ottaiano A, Franco R, Aiello TA et al (2006) Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II–III colorectal cancer patients. Clin Cancer Res 12:2795–2803
Kim J, Takeuchi H, Lam ST et al (2005) Chemokine receptor CXCR4 expression in colorectal cancer patients increases the risk for recurrence and for poor survival. J Clin Oncol 23:2744–2753
Schimanski CC, Schwald S, Simiantonaki N et al (2005) Effect of chemokine receptors CXCR4 and CCR7 on the metastatic behavior of human colorectal cancer. Clin Cancer Res 11:1743–1750
Ottaiano A, di Palma A, Napolitano M et al (2005) Inhibitory effects of anti-CXCR4 antibodies on human colon cancer cells. Cancer Immunol Immunother 54:781–791
Jordan NJ, Kolios G, Abbot SE et al (1999) Expression of functional CXCR4 chemokine receptors on human colonic epithelial cells. J Clin Invest 104:1061–1069
Dwinell MB, Eckmann L, Leopard JD et al (1999) Chemokine receptor expression by human intestinal epithelial cells. Gastroenterology 117:359–367
Rollins BJ (1997) Chemokines. Blood 90:909–928
Salvucci O, Bouchard A, Baccarelli A et al (2006) The role of CXCR4 receptor expression in breast cancer: a large tissue microarray study. Breast Cancer Res Treat 97:275–283
Wang N, Wu QL, Fang Y et al (2005) Expression of chemokine receptor CXCR4 in nasopharyngeal carcinoma: pattern of expression and correlation with clinical outcome. J Transl Med 3:26
Spano JP, Andre F, Morat L et al (2004) Chemokine receptor CXCR4 and early-stage non-small cell lung cancer: pattern of expression and correlation with outcome. Ann Oncol 15:613–617
Woo SU, Bae JW, Kim CH, et al (2007) A significant correlation between nuclear CXCR4 expression and axillary lymph node metastasis in hormonal receptor negative breast cancer. Ann Surg Oncol
Dierssen JW, de Miranda NF, Ferrone S et al (2007) HNPCC versus sporadic microsatellite-unstable colon cancers follow different routes toward loss of HLA class I expression. BMC Cancer 7:33
Speetjens FM, de Bruin EC, Morreau H et al (2008) Clinical impact of HLA class I expression in rectal cancer. Cancer Immunol Immunother 57:601–609
de Jong AE, van PM, Hendriks Y et al (2004) Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res 10:972–980
Balkwill F (2004) The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol 14:171–179
Contento RL, Molon B, Boularan C et al (2008) CXCR4-CCR5: a couple modulating T cell functions. Proc Natl Acad Sci U S A 105:10101–10106
Wald O, Izhar U, Amir G et al (2006) CD4+CXCR4highCD69+ T cells accumulate in lung adenocarcinoma. J Immunol 177:6983–6990
Shim H, Lau SK, Devi S et al (2006) Lower expression of CXCR4 in lymph node metastases than in primary breast cancers: potential regulation by ligand-dependent degradation and HIF-1alpha. Biochem Biophys Res Commun 346:252–258
Lin SY, Makino K, Xia W et al (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol 3:802–808
Huang YC, Hsiao YC, Chen YJ, et al (2007) Stromal cell-derived factor-1 enhances motility and integrin up-regulation through CXCR4, ERK and NF-kappaB-dependent pathway in human lung cancer cells. Biochem Pharmacol
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Speetjens, F.M., Liefers, G.J., Korbee, C.J. et al. Nuclear Localization of CXCR4 Determines Prognosis for Colorectal Cancer Patients. Cancer Microenvironment 2, 1–7 (2009). https://doi.org/10.1007/s12307-008-0016-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12307-008-0016-1