Abstract
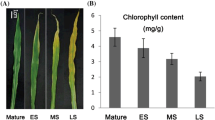
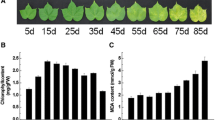
Red clover (Trifolium pratense L.) is an important cool-season legume plant, which is used as forage. Leaf senescence is a critical developmental process that negatively affects plant quality and yield. The regulatory mechanism of leaf senescence has been studied, and genes involved in leaf senescence have been cloned and characterized in many plants. However, those works mainly focused on model plants. Information about regulatory pathways and the genes involved in leaf senescence in red clover is very sparse. In this study, to better understand leaf senescence in red clover, transcriptome analysis of mature and senescent leaves was investigated using RNA-Seq. A total of about 35,067 genes were identified, and 481 genes were differentially expressed in mature and senescent leaves. Some identified differentially expressed genes showed similar expression patterns as those involved in leaf senescence in other species, such as Arabidopsis, Medicago truncatula and rice. Differentially expressed genes were confirmed by quantitative real-time PCR (qRT-PCR). Genes involved in signal transduction, transportation and metabolism of plant hormones, transcription factors and plant senescence were upregulated, while the downregulated genes were primarily involved in nutrient cycling, lipid/carbohydrate metabolism, hormone response and other processes. There were 64 differentially expressed transcription factor genes identified by RNA-Seq, including ERF, WRKY, bHLH, MYB and NAC. A total of 90 genes involved in biosynthesis, metabolism and transduction of plant hormones, including abscisic acid, jasmonic acid, cyokinin, brassinosteroid, salicylic acid and ethylene, were identified. Furthermore, 207 genes with direct roles in leaf senescence were demonstrated, such as senescence-associated genes. These genes were associated with senescence in other plants. Transcriptome analysis of mature and senescent leaves in red clover provides a large number of differentially expressed genes. Further analysis and identification of senescence-associated genes can provide new insight into the regulatory mechanisms of leaf development and senescence in legume plant and red clover.






Similar content being viewed by others
References
Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. https://doi.org/10.1093/bioinformatics/btu638
Balazadeh S, Riano-Pachon DM, Mueller-Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol 10(Suppl 1):63–75. https://doi.org/10.1111/j.1438-8677.2008.00088.x
Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B (2011) ORS1, an H(2)O(2)-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360. https://doi.org/10.1093/mp/ssq080
Bipfubusa M, Rocher S, Bertrand A, Castonguay Y, Renaut J (2016) Dataset of protein changes induced by cold acclimation in red clover (Trifolium pratense L.) populations recurrently selected for improved freezing tolerance. Data Brief 8:570–574. https://doi.org/10.1016/j.dib.2016.06.003
Buchanan-Wollaston V et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42:567–585. https://doi.org/10.1111/j.1365-313X.2005.02399.x
Burg SP (1968) Ethylene, plant senescence and abscission. Plant Physiol 43:1503–1511
Chakrabarti M, Dinkins RD, Hunt AG (2016) De novo transcriptome assembly and dynamic spatial gene expression analysis in red clover. Plant Genome. https://doi.org/10.3835/plantgenome2015.06.0048
Chao Y, Kang J, Zhang T, Yang Q, Gruber MY, Sun Y (2014) Disruption of the homogentisate solanesyltransferase gene results in albino and dwarf phenotypes and root, trichome and stomata defects in Arabidopsis thaliana. PLoS One 9:e94031. https://doi.org/10.1371/journal.pone.0094031
Chen Y, Qiu K, Kuai B, Ding Y (2011) Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Physiol Plant 142:361–371. https://doi.org/10.1111/j.1399-3054.2011.01472.x
Colquhoun AJ, Hillman JR (1972) The effects of abscisic acid on senescence in leaf discs of radish, Raphanus sativus L. Planta 105:213–224. https://doi.org/10.1007/BF00385393
De Michele R, Formentin E, Lo Schiavo F (2009a) Legume leaf senescence: a transcriptional analysis. Plant Signal Behav 4:319–320
De Michele R et al (2009b) Transcriptome analysis of Medicago truncatula leaf senescence: similarities and differences in metabolic and transcriptional regulations as compared with Arabidopsis, nodule senescence and nitric oxide signalling. New Phytol 181:563–575. https://doi.org/10.1111/j.1469-8137.2008.02684.x
De Vega JJ et al (2015) Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci Rep 5:17394. https://doi.org/10.1038/srep17394
Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C (1998) Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J 14:603–611
Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13:1025–1033
Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14:1557–1566
Flythe M, Kagan I (2010) Antimicrobial effect of red clover (Trifolium pratense) phenolic extract on the ruminal hyper ammonia-producing bacterium, Clostridium sticklandii. Curr Microbiol 61:125–131. https://doi.org/10.1007/s00284-010-9586-5
Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270:1986–1988
Gan S, Amasino RM (1997) Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol 113:313–319
Gregersen PL, Holm PB (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol J 5:192–206. https://doi.org/10.1111/j.1467-7652.2006.00232.x
Grover A, Sabat SC, Mohanty P (1986) Relative sensitivity of various spectral forms of photosynthetic pigments to leaf senescence in wheat (Triticum aestivum L.). Photosynth Res 10:223–231. https://doi.org/10.1007/BF00118287
Gunesekera B, Torabinejad J, Robinson J, Gillaspy GE (2007) Inositol polyphosphate 5-phosphatases 1 and 2 are required for regulating seedling growth. Plant Physiol 143:1408–1417. https://doi.org/10.1104/pp.106.089474
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612. https://doi.org/10.1111/j.1365-313X.2006.02723.x
Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27:521–549. https://doi.org/10.1111/j.1365-3040.2003.01158.x
Gut H, Rutz C, Matile P, Thomas H (1987) Leaf senescence in a non-yellowing mutant of Festuca pratensis: degradation of carotenoids. Physiol Plant 70:659–663. https://doi.org/10.1111/j.1399-3054.1987.tb04321.x
He Y, Gan S (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14:805–815
He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128:876–884. https://doi.org/10.1104/pp.010843
Hortensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53:927–937
Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53:927–937. https://doi.org/10.1093/jexbot/53.370.927
Hu HH, Jing CQ, Liu R, Li WD, Feng HG (2015) Cloning and transformation analysis of isoflavone synthase gene into Minshan Trifolium pratense. Genet Mol Res 14:9291–9297. https://doi.org/10.4238/2015.August.10.9
Istvanek J, Jaros M, Krenek A, Repkova J (2014) Genome assembly and annotation for red clover (Trifolium pratense; Fabaceae). Am J Bot 101:327–337. https://doi.org/10.3732/ajb.1300340
Jaradat MR, Feurtado JA, Huang D, Lu Y, Cutler AJ (2013) Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol 13:192. https://doi.org/10.1186/1471-2229-13-192
Jost R et al (2005) Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth Res 86:491–508. https://doi.org/10.1007/s11120-005-7386-8
Kant S, Burch D, Badenhorst P, Palanisamy R, Mason J, Spangenberg G (2015) Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLoS One 10:e0116349. https://doi.org/10.1371/journal.pone.0116349
Kaurinovic B, Popovic M, Vlaisavljevic S, Schwartsova H, Vojinovic-Miloradov M (2012) Antioxidant profile of Trifolium pratense L. Molecules 17:11156–11172. https://doi.org/10.3390/molecules170911156
Khan M, Rozhon W, Poppenberger B (2014) The role of hormones in the aging of plants—a mini-review. Gerontology 60:49–55. https://doi.org/10.1159/000354334
Kim JI, Murphy AS, Baek D, Lee SW, Yun DJ, Bressan RA, Narasimhan ML (2011) YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62:3981–3992. https://doi.org/10.1093/jxb/err094
Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC (2013) Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol 54:1660–1672. https://doi.org/10.1093/pcp/pct113
Kim J, Chang C, Tucker ML (2015) To grow old: regulatory role of ethylene and jasmonic acid in senescence. Front Plant Sci 6:20. https://doi.org/10.3389/fpls.2015.00020
Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141:1376–1388. https://doi.org/10.1104/pp.106.082941
Kong W, Li Y, Zhang M, Jin F, Li J (2015) A novel arabidopsis microRNA promotes IAA biosynthesis via the indole-3-acetaldoxime pathway by suppressing superroot1. Plant Cell Physiol 56:715–726. https://doi.org/10.1093/pcp/pcu216
Kou X, Watkins CB, Gan SS (2012) Arabidopsis AtNAP regulates fruit senescence. J Exp Bot 63:6139–6147. https://doi.org/10.1093/jxb/ers266
Kumar M, Singh VP, Arora A, Singh N (2014) The role of abscisic acid (ABA) in ethylene insensitive Gladiolus (Gladiolus grandiflora Hort.) flower senescence. Acta Physiol Plant 36:151–159. https://doi.org/10.1007/s11738-013-1395-6
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Li FF, Zhan D, Xu LX, Han LB, Zhang XZ (2014) Antioxidant and hormone responses to heat stress in two Kentucky Bluegrass cultivars contrasting in heat tolerance. J Am Soc Hortic Sci 139:587–596
Lichtenthaler HK (1987) Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In: Methods in Enzymology, vol 148. Academic Press, pp 350–382. http://dx.doi.org/10.1016/0076-6879(87)48036-1
Lin M, Pang C, Fan S, Song M, Wei H, Yu S (2015) Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol 15:43. https://doi.org/10.1186/s12870-015-0433-5
Liu J, Li J, Wang H, Fu Z, Liu J, Yu Y (2011) Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J Exp Bot 62:825–840. https://doi.org/10.1093/jxb/erq324
Lu P et al (2014) RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J 78:578–590. https://doi.org/10.1111/tpj.12494
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
Mayak S, Dilley DR (1976) Regulation of senescence in carnation (Dianthus caryophyllus): effect of abscisic acid and carbon dioxide on ethylene production. Plant Physiol 58:663–665
Mayak S, Halevy AH (1970) Cytokinin activity in rose petals and its relation to senescence. Plant Physiol 46:497–499
Mayak S, Halevy AH (1972) Interrelationships of ethylene and abscisic Acid in the control of rose petal senescence. Plant Physiol 50:341–346
Mikkelsen MD, Naur P, Halkier BA (2004) Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37:770–777
Mikkelsen MD, Fuller VL, Hansen BG, Nafisi M, Olsen CE, Nielsen HB, Halkier BA (2009) Controlled indole-3-acetaldoxime production through ethanol-induced expression of CYP79B2. Planta 229:1209–1217. https://doi.org/10.1007/s00425-009-0907-5
Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP (2011) Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol 156:1149–1163. https://doi.org/10.1104/pp.111.174805
Nam HG (1997) The molecular genetic analysis of leaf senescence. Curr Opin Biotechnol 8:200–207
Noh YS, Amasino RM (1999) Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol Biol 41:181–194
Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucl Acids Res 38:D822–D827. https://doi.org/10.1093/nar/gkp805
Rinerson CI et al (2015) The WRKY transcription factor family and senescence in switchgrass. BMC Genom 16:912. https://doi.org/10.1186/s12864-015-2057-4
Robatzek S, Somssich IE (2001) A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J 28:123–133
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Samet JS, Sinclair TR (1980) Leaf senescence and abscisic acid in leaves of field-grown soybean. Plant Physiol 66:1164–1168
Seltmann MA, Stingl NE, Lautenschlaeger JK, Krischke M, Mueller MJ, Berger S (2010) Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol 152:1940–1950. https://doi.org/10.1104/pp.110.153114
Sharabi-Schwager M, Lers A, Samach A, Guy CL, Porat R (2010) Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J Exp Bot 61:261–273. https://doi.org/10.1093/jxb/erp300
Sullivan ML, Quesenberry KH (2006) Red clover (Trifolium pratense). Methods Mol Biol 343:369–383. https://doi.org/10.1385/1-59745-130-4:369
Sullivan ML, Quesenberry KH (2015) Clover, red (Trifolium pratense). Methods Mol Biol 1223:237–254. https://doi.org/10.1007/978-1-4939-1695-5_19
Tang G-L, Li X-Y, Lin L-S, Zeng F-J, Gu Z-Y (2015) Girdling-induced Alhagi sparsifolia senescence and chlorophyll fluorescence changes. Photosynthetica 53:585–596. https://doi.org/10.1007/s11099-015-0148-8
Taylor NL, Quesenberry KH (1996) Red clover science. Current plant sciences and biology in agriculture. Kluwer, Dordrecht
Thompson JE, Mayak S, Shinitzky M, Halevy AH (1982) Acceleration of membrane senescence in cut carnation flowers by treatment with ethylene. Plant Physiol 69:859–863
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. https://doi.org/10.1093/bioinformatics/btp120
Trapnell C et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. https://doi.org/10.1038/nbt.1621
Wistrom C, Villeponteau B (1992) Cloning and expression of SAG: a novel marker of cellular senescence. Exp Cell Res 199:355–362
Woo HR et al (2016) Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in arabidopsis. Plant Physiol 171:452–467. https://doi.org/10.1104/pp.15.01929
Wu XY et al (2016) Transcriptome profiling of developmental leaf senescence in sorghum (Sorghum bicolor). Plant Mol Biol 92:555–580. https://doi.org/10.1007/s11103-016-0532-1
Xing S et al (2010) Disruption of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) gene results in albino, dwarf and defects in trichome initiation and stomata closure in Arabidopsis. Cell Res 20:688–700. https://doi.org/10.1038/cr.2010.54
Xu H, Wang X, Chen J (2010) Overexpression of the Rap2.4f transcriptional factor in Arabidopsis promotes leaf senescence. Sci China Life Sci 53:1221–1226. https://doi.org/10.1007/s11427-010-4068-3
Yang SD, Seo PJ, Yoon HK, Park CM (2011) The arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23:2155–2168. https://doi.org/10.1105/tpc.111.084913
Yates SA et al (2014) De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genom 15:453. https://doi.org/10.1186/1471-2164-15-453
Yeung KS, Gubili J (2008) Red clover (Trifolium pratense). J Soc Integr Oncol 6:176–177
Young AJ, Wellings R, Britton G (1991) The fate of chloroplast pigments during senescence of primary leaves of Hordeum vulgare and Avena sativum. J Plant Physiol 137:701–705. https://doi.org/10.1016/S0176-1617(11)81225-3
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Zhan J, He HY, Wang TJ, Wang AQ, Li CZ, He LF (2013) Aluminum-induced programmed cell death promoted by AhSAG, a senescence-associated gene in Arachis hypoganea L. Plant Sci 210:108–117. https://doi.org/10.1016/j.plantsci.2013.05.012
Zhang X, Ju HW, Chung MS, Huang P, Ahn SJ, Kim CS (2011) The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol 52:138–148. https://doi.org/10.1093/pcp/pcq180
Zhang K, Xia X, Zhang Y, Gan SS (2012) An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J 69:667–678. https://doi.org/10.1111/j.1365-313X.2011.04821.x
Zhao Y et al (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113:1949–1954. https://doi.org/10.1073/pnas.1522840113
Zwack PJ, Rashotte AM (2013) Cytokinin inhibition of leaf senescence. Plant Signal Behav 8:e24737. https://doi.org/10.4161/psb.24737
Acknowledgements
We thank Dr. Feifei Li from the Top Green Group for providing red clover seeds. The program was supported by the Fundamental Research Funds for the Central Universities (No. 2017ZY16), National Natural Science Foundation of China (Nos. 31672477 and 31601989) and Scientific Technology Plan Program of Shen Zhen (No. JCYJ20160331151245672).
Author information
Authors and Affiliations
Contributions
LH conceived and designed the research. YC, JY, TG and FL conducted experiments. YC and TG analyzed data. YC wrote the manuscript. LX provided suggestion and revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chao, Y., Xie, L., Yuan, J. et al. Transcriptome analysis of leaf senescence in red clover (Trifolium pratense L.). Physiol Mol Biol Plants 24, 753–765 (2018). https://doi.org/10.1007/s12298-018-0562-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-018-0562-z