Abstract
Background
We aimed to investigate the potential of microRNA expression profiles to predict survival in breast cancer.
Methods
MicroRNA and mRNA expression data of breast cancer were downloaded from The Cancer Genome Atlas. LASSO regression was used to identify microRNAs signature predicting survival of breast cancer patients. Transfection experiment was conducted to explore the influence of microRNAs on their potential targets.
Results
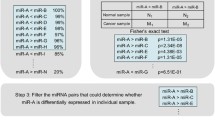
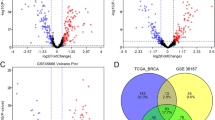
We identified 56 differentially expressed microRNAs in breast cancer tissues compared to adjacent normal tissues. 10 microRNAs with non-zero coefficient were selected from the 56 microRNAs using LASSO Cox regression. After predicting the targets for the 10 microRNAs, we further obtained 155 targets that were associated with overall survival of breast cancer patients. Spearman’s correlation analysis found that the expression of SCUBE2, SCRN3, YTHDF3, ITFG1, ITPRIPL2, and JAK1 was an inversely correlated with their microRNAs. Transfection experiment showed that YTHDF3 was down-regulated in cells transfected with miR-106b-5p mimics compared with those transfected with negative control of mimics (fold change 4.21; P < 0.01).
Conclusions
In conclusion, we identified a 10-miRNA signature associated with prognosis of breast cancer patients. The expression of YTHDF3 was down-regulated by miR-106b-5p.





Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017; 67:7–30
Esbah O, Oksuzoglu B. Prognostic & predictive factors for planning adjuvant chemotherapy of early-stage breast cancer. Indian J Med Res. 2017;146:563–71.
Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin Cancer Biol. 2018;52:56–73.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31.
Mandujano-Tinoco EA, Garcia-Venzor A, Melendez-Zajgla J, Maldonado V. New emerging roles of microRNAs in breast cancer. Breast Cancer Res Treat. 2018;171:247–59
Teoh SL, Das S. The role of MicroRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr Pharm Des. 2017;23:1845–59.
Tang Y, Zhou X, Ji J, Chen L, Cao J, Luo J, et al. High expression levels of miR-21 and miR-210 predict unfavorable survival in breast cancer: a systemic review and meta-analysis. Int J Biol Mark. 2015;30:e347-58.
Kim SY, Kawaguchi T, Yan L, Young J, Qi Q, Takabe K. Clinical relevance of microRNA expressions in breast cancer validated using The Cancer Genome Atlas (TCGA). Ann Surg Oncol. 2017;24:2943–9.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B (Methodol). 1996;58:267–88.
Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19:A68–77.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84.
Alencar AJ, Malumbres R, Kozloski GA, Advani R, Talreja N, Chinichian S, et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res. 2011;17:4125–35.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005
Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146-52.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Zhong S, Wang J, Hou J, Zhang Q, Xu H, Hu J, et al. Circular RNA hsa_circ_0000993 inhibits metastasis of gastric cancer cells. Epigenomics. 2018;10:1301–13.
Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–306.
Hayes J, Thygesen H, Tumilson C, Droop A, Boissinot M, Hughes TA, et al. Prediction of clinical outcome in glioblastoma using a biologically relevant nine-microRNA signature. Mol Oncol. 2015;9:704–14.
Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5:395–402.
Barbano R, Pasculli B, Rendina M, Fontana A, Fusilli C, Copetti M, et al. Stepwise analysis of MIR9 loci identifies miR-9-5p to be involved in Oestrogen regulated pathways in breast cancer patients. Sci Rep. 2017;7:45283.
Jiang CF, Shi ZM, Li DM, Qian YC, Ren Y, Bai XM, et al. Estrogen-induced miR-196a elevation promotes tumor growth and metastasis via targeting SPRED1 in breast cancer. Mol Cancer. 2018;17:83.
Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587–95.
Hu Y, Zhu Q, Tang L. MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS One. 2014;9:e92099.
Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X, et al. Downregulation of miR-106b induced breast cancer cell invasion and motility in association with overexpression of matrix metalloproteinase 2. Cancer Sci. 2014;105:18–25.
Lin YC, Chen CC, Cheng CJ, Yang RB. Domain and functional analysis of a novel breast tumor suppressor protein, SCUBE2. J Biol Chem. 2011;286:27039–47.
Lin YC, Lee YC, Li LH, Cheng CJ, Yang RB. Tumor suppressor SCUBE2 inhibits breast-cancer cell migration and invasion through the reversal of epithelial-mesenchymal transition. J Cell Sci. 2014;127:85–100.
Cheng CJ, Lin YC, Tsai MT, Chen CS, Hsieh MC, Chen CL, et al. SCUBE2 suppresses breast tumor cell proliferation and confers a favorable prognosis in invasive breast cancer. Cancer Res. 2009;69:3634–41.
Fan W, Xie J, Xia J, Zhang Y, Yang M, Wang H, et al. RUVBL1-ITFG1 interaction is required for collective invasion in breast cancer. Biochim Biophys Acta. 2017;1861:1788–800.
Quintas-Cardama A, Verstovsek S. Molecular pathways: Jak/STAT pathway: mutations, inhibitors, and resistance. Clin Cancer Res. 2013;19:1933–40.
Lee HJ, Seo NJ, Jeong SJ, Park Y, Jung DB, Koh W, et al. Oral administration of penta-O-galloyl-beta-d-glucose suppresses triple-negative breast cancer xenograft growth and metastasis in strong association with JAK1-STAT3 inhibition. Carcinogenesis. 2011;32:804–11.
Kaushik N, Kim MJ, Kim RK, Kumar Kaushik N, Seong KM, Nam SY, et al. Low-dose radiation decreases tumor progression via the inhibition of the JAK1/STAT3 signaling axis in breast cancer cell lines. Sci Rep. 2017;7:43361.
Funding
This study was funded by the National Natural Science Foundation of China (Grant number 81602551) and the young talents program of Jiangsu Cancer Hospital (2017YQL-10).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12282_2018_926_MOESM1_ESM.tif
Figure S1 Overall survival analysis of breast cancer patients with high-expression genes and low-expression genes. The number following “=” is the median survival time for each group (509 patients for each group) (TIF 561 KB)
About this article
Cite this article
Liu, M., Zhou, S., Wang, J. et al. Identification of genes associated with survival of breast cancer patients. Breast Cancer 26, 317–325 (2019). https://doi.org/10.1007/s12282-018-0926-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-018-0926-9