Abstract
Background
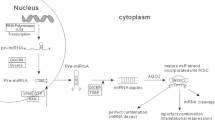
MicroRNAs constitute a large family of non-coding RNAs, which actively participate in tumorigenesis by regulating a set of mRNAs of distinct signaling pathways. An altered expression of these molecules has been found in different tumorigenic processes of breast cancer, the most common type of cancer in the female population worldwide.
Purpose
The objective of this review is to discuss how miRNAs become master regulators in breast tumorigenesis.
Methods
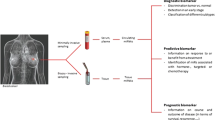
An integrative review of miRNAs and breast cancer literature from the last 5 years was done on PubMed. We summarize recent works showing that the defects on the biogenesis of miRNAs are associated with different breast cancer characteristics. Then, we show several examples that demonstrate the link between cellular processes regulated by miRNAs and the hallmarks of breast cancer. Finally, we examine the complexity in the regulation of these molecules as they are modulated by other non-coding RNAs and the clinical applications of miRNAs as they could serve as good diagnostic and classification tools.
Conclusion
The information presented in this review is important to encourage new directed studies that consider microRNAs as a good tool to improve the diagnostic and treatment alternatives in breast cancer.



Similar content being viewed by others
Abbreviations
- BC:
-
Breast cancer
- TNBC:
-
Triple-negative breast cancer
- EMT:
-
Epithelial-to-mesenchymal transition
- BCSC:
-
Breast cancer stem cells
- EGR1:
-
Early growth response protein 1
- SOCS1:
-
Suppressor of cytokine signaling 1
- CDKN1B:
-
Cyclin-dependent kinase inhibitor 1B
- STAT5A:
-
Signal transducer and activator of transcription 5a
- PTEN:
-
Phosphatase and tensin homolog
- Akt:
-
Protein kinase B
- Bcl-w:
-
Bcl-2-like protein 2
- ERα:
-
Estrogen receptor alpha
- E2F3:
-
E2F Transcription factor 3
- SMAD7:
-
Mothers against decapentaplegic homolog 7
- CDK8:
-
Cyclin-dependent kinase 8
- BNDF:
-
Brain-derived neurotrophic factor
- SIAH1:
-
Siah E3 ubiquitin protein ligase 1
- MCL-1:
-
Induced myeloid leukemia cell differentiation protein
- ADIPOR1:
-
Adiponectin receptor 1
- TRPS1:
-
Transcriptional repressor GATA binding 1
- ADAM-17:
-
ADAM metallopeptidase domain 17
- uPAR7b:
-
uPAR isoform 2
- NIM:
-
N-Myc interactor
- NOV/CCN3:
-
Nephroblastoma overexpressed
- ERBB3:
-
Erb-B2 receptor tyrosine kinase 3
- Kras:
-
Proto-oncogene for Kirsten rat sarcoma viral oncogene
- MMP9:
-
Matrix metallopeptidase 9
- CXCR4:
-
C-X-C chemokine receptor type 4
- ZBTB10:
-
Zinc finger and BTB domain containing 10
- Ang2:
-
Angiopoietin-2
- IL8:
-
Interleukin 8
- CXCL1:
-
Chemokine (C-X-C motif) ligand 1
- VHL:
-
Von Hippel–Lindau tumor suppressor
- hTERT:
-
Human telomerase reverse transcriptase
- C/EBPβ:
-
CCAAT/enhancer-binding protein beta
- LDHA:
-
Lactate dehydrogenase A
- PK:
-
Pyruvate kinase
- ERRγ:
-
Estrogen-related receptor gamma
- GABPA:
-
GA-binding protein alpha
- ISCU:
-
Iron–sulfur cluster assembly enzyme
- MiCB:
-
MHC class I polypeptide-related sequence B
- TERF1:
-
Telomeric repeat-binding factor 1
- FOXO3a:
-
Forkhead box O3
- BRCA1:
-
Breast cancer type 1 susceptibility protein
- NFKB:
-
Nuclear factor kappa B
- OxPhos:
-
Oxidative phosphorylation
References
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65(16):7065–7070
Dai X, Xiang L, Li T, Bai Z (2016) Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J Cancer 7(10):1281–1294
Kim VN, Nam JW (2006) Genomics of microRNA. Trends Genet 22(3):165–173
Borchert GM, Lanier W, Davidson BL (2006) RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13(12):1097–1101
Bordinhao AL, Evangelista AF, Oliveira RJ, Macedo T, Silveira HC, Reis RM, Marques MM (2016) MicroRNA profiling in human breast cancer cell lines exposed to the anti-neoplastic drug cediranib. Oncol Rep 36(6):3197–3206
Koo CX, Kobiyama K, Shen YJ, LeBert N, Ahmad S, Khatoo M, Aoshi T, Gasser S, Ishii KJ (2015) RNA polymerase III regulates cytosolic RNA:DNA hybrids and intracellular microRNA expression. J Biol Chem 290(12):7463–7473
Poursadegh Zonouzi AA, Shekari M, Nejatizadeh A, Shakerizadeh S, Fardmanesh H, Poursadegh Zonouzi A, Rahmati-Yamchi M, Tozihi M (2017) Impaired expression of Drosha in breast cancer. Breast Dis 37:55
Fardmanesh H, Shekari M, Movafagh A, Shargh SA, Zonouzi AAP, Shakerizadeh S, Zonouzi AP, Hosseinzadeh A (2016) Upregulation of the double-stranded RNA binding protein DGCR8 in invasive ductal breast carcinoma. Gene 581(2):146–151
Dedes KJ, Natrajan R, Lambros MB, Geyer FC, Lopez-Garcia MA, Savage K, Jones RL, Reis-Filho JS (2011) Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer 47(1):138–150
Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G et al (2010) A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 18(4):303–315
Vaidyanathan S, Thangavelu PU, Duijf PH (2016) Overexpression of Ran GTPase components regulating nuclear export, but not mitotic spindle assembly, marks chromosome instability and poor prognosis in breast cancer. Target Oncol 11(5):677–686
Kwon SY, Lee JH, Kim B, Park JW, Kwon TK, Kang SH, Kim S (2014) Complexity in regulation of microRNA machinery components in invasive breast carcinoma. Pathol Oncol Res 20(3):697–705
Spoelstra NS, Cittelly DM, Christenson JL, Gordon MA, Elias A, Jedlicka P, Richer JK (2016) Dicer expression in estrogen receptor-positive versus triple-negative breast cancer: an antibody comparison. Hum Pathol 56:40–51
Sha LY, Zhang Y, Wang W, Sui X, Liu SK, Wang T, Zhang H (2016) MiR-18a upregulation decreases Dicer expression and confers paclitaxel resistance in triple negative breast cancer. Eur Rev Med Pharmacol Sci 20(11):2201–2208
Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA et al (2014) Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26(5):707–721
Breving K, Esquela-Kerscher A (2010) The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol 42(8):1316–1329
Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC (2011) Widespread regulatory activity of vertebrate microRNA* species. RNA 17(2):312–326
Rhodes LV, Martin EC, Segar HC, Miller DF, Buechlein A, Rusch DB, Nephew KP, Burow ME, Collins-Burow BM (2015) Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in triple-negative breast cancer. Oncotarget 6(18):16638–16652
Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD (2009) New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19(7):1175–1183
Oliveto S, Mancino M, Manfrini N, Biffo S (2017) Role of microRNAs in translation regulation and cancer. World J Biol Chem 8(1):45–56
Chapat C, Jafarnejad SM, Matta-Camacho E, Hesketh GG, Gelbart IA, Attig J, Gkogkas CG, Alain T, Stern-Ginossar N, Fabian MR et al (2017) Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc Natl Acad Sci USA 114(21):5425–5430
Ramchandran R, Chaluvally-Raghavan P (2017) miRNA-mediated RNA activation in mammalian cells. Adv Exp Med Biol 983:81–89
Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S (2007) The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res 67(22):11001–11011
Zhu S, Si ML, Wu H, Mo YY (2007) MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 282(19):14328–14336
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J et al (2007) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131(6):1109–1123
Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO et al (2007) MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 8(10):R214
Liang Z, Wu H, Reddy S, Zhu A, Wang S, Blevins D, Yoon Y, Zhang Y, Shim H (2007) Blockade of invasion and metastasis of breast cancer cells via targeting CXCR4 with an artificial microRNA. Biochem Biophys Res Commun 363(3):542–546
Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J (2008) hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14(5):1340–1348
Zhu W, Qin W, Atasoy U, Sauter ER (2009) Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes 2:89
Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ (2010) Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg 251(3):499–505
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, Cheung DG, Bolon B, Anderlucci L, Alder H et al (2013) Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet 9(3):e1003311
Li B, Lu Y, Wang H, Han X, Mao J, Li J, Yu L, Wang B, Fan S, Yu X et al (2016) miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother 79:93–101
Dentelli P, Traversa M, Rosso A, Togliatto G, Olgasi C, Marchio C, Provero P, Lembo A, Bon G, Annaratone L et al (2014) miR-221/222 control luminal breast cancer tumor progression by regulating different targets. Cell Cycle 13(11):1811–1826
Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, Li L, Qin J, Chen Y, Cho WC et al (2014) miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules 19(6):7122–7137
Han L, Liu B, Jiang L, Liu J, Han S (2016) MicroRNA-497 downregulation contributes to cell proliferation, migration, and invasion of estrogen receptor alpha negative breast cancer by targeting estrogen-related receptor alpha. Tumour Biol 37(10):13205–13214
Shen L, Li J, Xu L, Ma J, Li H, Xiao X, Zhao J, Fang L (2012) miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med 3(3):475–480
Zhang Y, Zhang Z, Li Z, Gong D, Zhan B, Man X, Kong C (2016) MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3. Oncol Rep 36(3):1293–1300
Liu J, Zhou Y, Shi Z, Hu Y, Meng T, Zhang X, Zhang S, Zhang J (2016) microRNA-497 modulates breast cancer cell proliferation, invasion, and survival by targeting SMAD7. DNA Cell Biol 35(9):521–529
Li XY, Luo QF, Wei CK, Li DF, Li J, Fang L (2014) MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int J Clin Exp Med 7(1):32–40
Zhang L, Ma P, Sun LM, Han YC, Li BL, Mi XY, Wang EH, Song M (2016) MiR-107 down-regulates SIAH1 expression in human breast cancer cells and silencing of miR-107 inhibits tumor growth in a nude mouse model of triple-negative breast cancer. Mol Carcinog 55(5):768–777
Gao B, Hao S, Tian W, Jiang Y, Zhang M, Guo L, Zhao J, Zhang G, Yan J, Luo D (2016) MicroRNA-107 is downregulated and having tumor suppressive effect in breast cancer by negatively regulating BDNF. J Gene Med. https://doi.org/10.1002/jgm.2932
Gao J, Li L, Wu M, Liu M, Xie X, Guo J, Tang H, Xie X (2013) MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS ONE 8(6):e65138
Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T et al (2011) TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal 4(177):ra41
Hwang MS, Yu N, Stinson SY, Yue P, Newman RJ, Allan BB, Dornan D (2013) miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS ONE 8(6):e66502
Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang L, Yang A, Zhao J, Jia L (2014) MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep 47(5):268–273
Falkenberg N, Anastasov N, Schaub A, Radulovic V, Schmitt M, Magdolen V, Aubele M: Secreted uPAR isoform 2 (uPAR7b) is a novel direct target of miR-221 (2015) Oncotarget 6(10):8103–8114
Rostas JW 3rd, Pruitt HC, Metge BJ, Mitra A, Bailey SK, Bae S, Singh KP, Devine DJ, Dyess DL, Richards WO et al (2014) microRNA-29 negatively regulates EMT regulator N-myc interactor in breast cancer. Mol Cancer 13:200
Dobson JR, Taipaleenmaki H, Hu YJ, Hong D, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Pratap J (2014) hsa-mir-30c promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/CCN3. Cancer Cell Int 14:73
Tavanafar F, Safaralizadeh R, Hosseinpour-Feizi MA, Mansoori B, Shanehbandi D, Mohammadi A, Baradaran B (2017) Restoration of miR-143 expression could inhibit migration and growth of MDA-MB-468 cells through down-regulating the expression of invasion-related factors. Biomed Pharmacother 91:920–924
Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, Liu M, Gao X, Liu Y, Zhao C et al (2014) miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer 13:220
Tang W, Yu F, Yao H, Cui X, Jiao Y, Lin L, Chen J, Yin D, Song E, Liu Q (2014) miR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene 33(20):2629–2638
He T, Qi F, Jia L, Wang S, Song N, Guo L, Fu Y, Luo Y (2014) MicroRNA-542-3p inhibits tumour angiogenesis by targeting angiopoietin-2. J Pathol 232(5):499–508
Jones R, Watson K, Bruce A, Nersesian S, Kitz J, Moorehead R (2017) Re-expression of miR-200c suppresses proliferation, colony formation and in vivo tumor growth of murine claudin-low mammary tumor cells. Oncotarget 8(14):23727–23749
Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C et al (2013) Tumour angiogenesis regulation by the miR-200 family. Nat Commun 4:2427
Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY et al (2014) Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 33(6):679–689
Hrdlickova R, Nehyba J, Bargmann W, Bose HR Jr (2014) Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS ONE 9(2):e86990
Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, Li B, Li Y, Li D, Wang ED et al (2012) A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J 31(8):1985–1998
Xiao X, Huang X, Ye F, Chen B, Song C, Wen J, Zhang Z, Zheng G, Tang H, Xie X (2016) The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci Rep 6:21735
Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, Chow A, O’Connor ST, Li S, Chin AR et al (2015) Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol 17(2):183–194
Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, Giguere V (2010) miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab 12(4):352–361
Serguienko A, Grad I, Wennerstrom AB, Meza-Zepeda LA, Thiede B, Stratford EW, Myklebost O, Munthe E (2015) Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA Oncotarget 6(4):2451–2465
Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL et al (2010) MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS ONE 5(4):e10345
Tsukerman P, Stern-Ginossar N, Gur C, Glasner A, Nachmani D, Bauman Y, Yamin R, Vitenshtein A, Stanietsky N, Bar-Mag T et al (2012) MiR-10b downregulates the stress-induced cell surface molecule MICB, a critical ligand for cancer cell recognition by natural killer cells. Cancer Res 72(21):5463–5472
Shen J, Pan J, Du C, Si W, Yao M, Xu L, Zheng H, Xu M, Chen D, Wang S et al (2017) Silencing NKG2D ligand-targeting miRNAs enhances natural killer cell-mediated cytotoxicity in breast cancer. Cell Death Dis 8(4):e2740
Elias S, Mandelboim O (2012) Battle of the midgets: innate microRNA networking. RNA Biol 9(6):792–798
Dinami R, Ercolani C, Petti E, Piazza S, Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R et al (2014) miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res 74(15):4145–4156
Czochor JR, Sulkowski P, Glazer PM (2016) miR-155 overexpression promotes genomic instability by reducing high-fidelity polymerase delta expression and activating error-prone DSB repair. Mol Cancer Res 14(4):363–373
Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM et al (2011) miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 41(2):210–220
Choi YE, Pan Y, Park E, Konstantinopoulos P, De S, D’Andrea A, Chowdhury D (2014) MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. Elife 3:e02445
Moazzeni H, Najafi A, Khani M (2017) Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Mol Cell Probes 34:45
Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X (2010) Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res 70(18):7176–7186
Xiang M, Birkbak NJ, Vafaizadeh V, Walker SR, Yeh JE, Liu S, Kroll Y, Boldin M, Taganov K, Groner B et al (2014) STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-kappaB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci Signal 7(310):ra11
Bahena-Ocampo I, Espinosa M, Ceballos-Cancino G, Lizarraga F, Campos-Arroyo D, Schwarz A, Garcia-Lopez P, Maldonado V, Melendez-Zajgla J (2016) miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep 17(7):1081
Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61(5):759–767
Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, Fang L (2013) MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int 13(1):95
Mandujano-Tinoco EA, Garcia-Venzor A, Munoz-Galindo L, Lizarraga-Sanchez F, Favela-Orozco A, Chavez-Gutierrez E, Krotzsch E, Salgado RM, Melendez-Zajgla J, Maldonado V:(2017) miRNA expression profile in multicellular breast cancer spheroids. Biochim Biophys Acta 1864:1642
Pan Y, Li J, Zhang Y, Wang N, Liang H, Liu Y, Zhang CY, Zen K, Gu H (2016) Slug-upregulated miR-221 promotes breast cancer progression through suppressing E-cadherin expression. Sci Rep 6:25798
Gao S, Wang Y, Wang M, Li Z, Zhao Z, Wang RX, Wu R, Yuan Z, Cui R, Jiao K et al (2017) MicroRNA-155, induced by FOXP3 through transcriptional repression of BRCA1, is associated with tumor initiation in human breast cancer. Oncotarget 8:41451
Ivancich M, Schrank Z, Wojdyla L, Leviskas B, Kuckovic A, Sanjali A, Puri N (2017) Treating cancer by targeting telomeres and telomerase. Antioxidants 6(1):E15
Bacci M, Giannoni E, Fearns A, Ribas R, Gao Q, Taddei ML, Pintus G, Dowsett M, Isacke CM, Martin LA et al (2016) miR-155 drives metabolic reprogramming of ER+ breast cancer cells following long-term estrogen deprivation and predicts clinical response to aromatase inhibitors Cancer Res 76(6):1615–1626
Lei K, Du W, Lin S, Yang L, Xu Y, Gao Y, Xu B, Tan S, Xu Y, Qian X et al (2016) 3B, a novel photosensitizer, inhibits glycolysis and inflammation via miR-155-5p and breaks the JAK/STAT3/SOCS1 feedback loop in human breast cancer cells. Biomed Pharmacother 82:141–150
Williams M, Cheng YY, Blenkiron C, Reid G:(2016) Exploring mechanisms of MicroRNA downregulation in cancer. Microrna 6:2
Vincent K, Pichler M, Lee GW, Ling H (2014) MicroRNAs, genomic instability and cancer. Int J Mol Sci 15(8):14475–14491
Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR (2010) Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol 4(3):255–266
Berton S, Cusan M, Segatto I, Citron F, D’Andrea S, Benevol S, Avanzo M, Dall’Acqua A, Schiappacassi M, Bristow RG et al (2017) Loss of p27kip1 increases genomic instability and induces radio-resistance in luminal breast cancer cells. Sci Rep 7(1):595
Selcuklu SD, Yakicier MC, Erson AE (2009) An investigation of microRNAs mapping to breast cancer related genomic gain and loss regions. Cancer Genet Cytogenet 189(1):15–23
Duru N, Gernapudi R, Eades G, Eckert R, Zhou Q (2015) Epigenetic regulation of miRNAs and breast cancer stem cells. Curr Pharmacol Rep 1(3):161–169
Sun H, Ding C, Zhang H, Gao J (2016) Let7 miRNAs sensitize breast cancer stem cells to radiation-induced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol Med Rep 14(4):3285–3292
Andorfer CA, Necela BM, Thompson EA, Perez EA (2011) MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med 17(6):313–319
Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC, Ding WQ (2016) Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res 18(1):90
Liu B, Su F, Chen M, Li Y, Qi X, Xiao J, Li X, Liu X, Liang W, Zhang Y et al (2017) Serum miR-21 and miR-125b as markers predicting neoadjuvant chemotherapy response and prognosis in stage II/III breast cancer. Hum Pathol 64:44
Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, Zhang GQ (2017) A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res 92(2):55–66
Mangolini A, Ferracin M, Zanzi MV, Saccenti E, Ebnaof SO, Poma VV, Sanz JM, Passaro A, Pedriali M, Frassoldati A et al (2015) Diagnostic and prognostic microRNAs in the serum of breast cancer patients measured by droplet digital PCR. Biomark Res 3:12
Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Horiguchi J (2017) Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet 62(1):15–24
Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL et al (2014) microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 5(5):1174–1184
Fkih MI, Privat M, Trimeche M, Penault-Llorca F, Bignon YJ, Kenani A miR-10b, miR-26a, miR-146a and miR-153 expression in triple negative vs non triple negative breast cancer: potential biomarkers. Pathol Oncol Res 23:815
Tsang JY, Ni YB, Ng EK, Shin VY, Mak KF, Go EM, Tawasil J, Chan SK, Ko CW, Kwong A et al (2015) MicroRNAs are differentially deregulated in mammary malignant phyllodes tumour. Histopathology 67(3):294–305
Raychaudhuri M, Bronger H, Buchner T, Kiechle M, Weichert W, Avril S (2017) MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat 162(3):511–521
Halvorsen AR, Helland A, Gromov P, Wielenga VT, Talman MM, Brunner N, Sandhu V, Borresen-Dale AL, Gromova I, Haakensen VD (2017) Profiling of microRNAs in tumor interstitial fluid of breast tumors - a novel resource to identify biomarkers for prognostic classification and detection of cancer. Mol Oncol 11(2):220–234
Das S, Lin TS (2016) The role of MicroRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr Pharm Des 23:1845
Radulovic V, Heider T, Richter S, Moertl S, Atkinson MJ, Anastasov N (2017) Differential response of normal and transformed mammary epithelial cells to combined treatment of anti-miR-21 and radiation. Int J Radiat Biol 93(4):361–372
Zhao D, Tu Y, Wan L, Bu L, Huang T, Sun X, Wang K, Shen B (2013) In vivo monitoring of angiogenesis inhibition via down-regulation of mir-21 in a VEGFR2-luc murine breast cancer model using bioluminescent imaging. PLoS ONE 8(8):e71472
Zhong S, Li W, Chen Z, Xu J, Zhao J (2013) MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene 531(1):8–14
Shen R, Wang Y, Wang CX, Yin M, Liu HL, Chen JP, Han JQ, Wang WB (2015) MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am J Transl Res 7(10):2115–2126
Zhao G, Li Y, Wang T (2017) Potentiation of docetaxel sensitivity by miR-638 via regulation of STARD10 pathway in human breast cancer cells. Biochem Biophys Res Commun 487(2):255–261
Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q (2014) MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene 33(24):3119–3128
Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou X, Zhang ZJ, Peng YH, Yang YZ, Yun JP (2014) miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis 35(2):469–478
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16(3):203–222
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17(5):272–283
Welch JD, Baran-Gale J, Perou CM, Sethupathy P, Prins JF (2015) Pseudogenes transcribed in breast invasive carcinoma show subtype-specific expression and ceRNA potential. BMC Genom 16:113
Rutnam ZJ, Du WW, Yang W, Yang X, Yang BB (2014) The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat Commun 5:2914
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J (2014) LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 5:e1287
Huan J, Xing L, Lin Q, Xui H, Qin X (2017) Long noncoding RNA CRNDE activates Wnt/beta-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am J Transl Res 9(4):1977–1989
Xu S, Kong D, Chen Q, Ping Y, Pang D (2017) Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer 16(1):129
Author information
Authors and Affiliations
Contributions
EAMT drafted the manuscript and edited the figures. AGV, JMZ, and VML helped to edit and review the manuscript. All authors read and approved the final version of the article.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Mandujano-Tinoco, E.A., García-Venzor, A., Melendez-Zajgla, J. et al. New emerging roles of microRNAs in breast cancer. Breast Cancer Res Treat 171, 247–259 (2018). https://doi.org/10.1007/s10549-018-4850-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-018-4850-7