Abstract
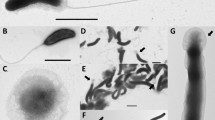
Three novel bacterial strains, HDW9AT, HDW9BT, and HDW9CT, isolated from the intestine of the diving beetles Cybister lewisianus and Cybister brevis, were characterized as three novel species using a polyphasic approach. The isolates were Gram-staining-positive, strictly aerobic, non-motile, and rod-shaped. They grew optimally at 30°C (pH 7) in the presence of 0.5% (wt/vol) NaCl. Phylogenetic analysis based on the 16S rRNA gene sequences revealed that they belong to the genus Leucobacter and are closely related to L. denitrificans M1T8B10T (98.4–98.7% sequence similarity). Average nucleotide identity (ANI) values among the isolates were 76.4–84.1%. ANI values for the isolates and the closest taxonomic species, L. denitrificans KACC 14055T, were 72.3–73.1%. The isolates showed ANI values of < 76.5% with all analyzable Leucobacter strains in the EzBioCloud database. The genomic DNA G + C content of the isolates was 60.3–62.5%. The polar lipid components were phosphatidylglycerol, diphosphatidylglycerol, and other unidentified glycolipids, phospholipids, and lipids. The major cellular fatty acids were anteiso-C15:0, iso-C16:0, and anteiso-C17:0. MK-10 was the major respiratory quinone, and MK-7 and MK-11 were the minor respiratory quinones. The whole-cell sugar components of the isolates were ribose, glucose, galactose, and mannose. The isolates harbored l-2,4-diaminobutyric acid, l-serine, l-lysine, l-aspartic acid, glycine, and d-glutamic acid within the cell wall peptidoglycan. Based on phylogenetic, phenotypic, chemotaxonomic, and genotypic analyses, strains HDW9AT, HDW9BT, and HDW9CT represent three novel species within the genus Leucobacter. We propose the name Leucobacter coleopterorum sp. nov. for strain HDW9AT (= KACC 21331T = KCTC 49317T = JCM 33667T), the name Leucobacter insecticola sp. nov. for strain HDW9BT (= KACC 21332T = KCTC 49318T = JCM 33668T), and the name Leucobacter viscericola sp. nov. for strain HDW9CT (= KACC 21333T = KCTC 49319T = JCM 33669T).
Similar content being viewed by others
References
Benga, L., Spröer, C., Schumann, P., Verbarg, S., Bunk, B., Engelhardt, E., Benten, W.P.M., and Sager, M. 2019. Leucobacter muris sp. nov., isolated from the nose of a laboratory mouse. Int. J. Syst. Evol. Microbiol. 69, 2095–2100.
Benson, H. 1994. Microbiological application. Laboratory manual in General Microbiology. Wan C. Publishers, Dubuque, USA.
Bousfield, G.R., Sugino, H., and Ward, D.N. 1985. Demonstration of a COOH-terminal extension on equine lutropin by means of a common acid-labile bond in equine lutropin and equine chorionic gonadotropin. J. Biol. Chem. 260, 9531–9533.
Chakraborty, S., Rene, E.R., and Lens, P.N.L. 2019. Reduction of selenite to elemental Se(0) with simultaneous degradation of phenol by co-cultures of Phanerochaete chrysosporium and Delftia lacustris. J. Microbiol. 57, 738–747.
Chen, I.M.A., Chu, K., Palaniappan, K., Pillay, M., Ratner, A., Huang, J., Huntemann, M., Varghese, N., White, J.R., Seshadri, R., et al. 2019. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677.
Chun, B.H., Lee, H.J., Jeong, S.E., Schumann, P., and Jeon, C.O. 2017. Leucobacter ruminantium sp. nov., isolated from the bovine rumen. Int. J. Syst. Evol. Microbiol. 67, 2634–2639.
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D.R., da Costa, M.S., Rooney, A.P., Yi, H., Xu, X.W., De Meyer, S., et al. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466.
Collins, M.D. and Jones, D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45, 316–354.
Contreras-Moreira, B. and Vinuesa, P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701.
Das, S., Dash, H.R., and Chakraborty, J. 2016. Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 100, 2967–2984.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Fitch, W.M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416.
Goszczynska, T. and Serfontein, J. 1998. Milk-Tween agar, a semiselective medium for isolation and differentiation of Pseudomonas syringae pv. syringae, Pseudomonas syringae pv. phaseolicola and Xanthomonas axonopodis pv. phaseoli. J. Microbiol. Methods 32, 65–72.
Ha, S.M., Kim, C.K., Roh, J., Byun, J.H., Yang, S.J., Choi, S.B., Chun, J., and Yong, D. 2019. Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann. Lab. Med. 39, 530–536.
Hiraishi, A., Ueda, Y., Ishihara, J., and Mori, T. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 42, 457–469.
Hou, S.Y., Wang, F., Zhang, M.Y., Cheng, J., and Zhang, Y.X. 2018. A novel pigmented and heavy metal biosorptive bacterium, Leucobacter epilobiisoli sp. nov., isolated from rhizosphere soil of Epilobium hirsutum L. Antonie van Leeuwenhoek 111, 1825–1833.
Kang, C.H., Kwon, Y.J., and So, J.S. 2016. Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 89, 64–69.
Kumar, S., Stecher, G., and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Lai, W.A., Lin, S.Y., Hameed, A., Hsu, Y.H., Liu, Y.C., Huang, H.R., Shen, F.T., and Young, C.C. 2015. Leucobacter zeae sp. nov., isolated from the rhizosphere of maize (Zea mays L.). Int. J. Syst. Evol. Microbiol. 65, 4734–4742.
Lane, D. 1991. 16S/23S rRNA sequencing. In Stackbrandt, E. and Goodfellow, M. (eds.), Nucleic acid techniques in bacterial systematics, pp. 115–175. John Wiley & Sons Ltd., Hoboken, New Jersey, USA.
Lee, I., Kim, Y.O., Park, S.C., and Chun, J. 2016. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103.
Li, H., Huang, S., and Zhang, Y. 2016. Cr(VI) removal from aqueous solution by thermophilic denitrifying bacterium Chelatococcus daeguensis TAD1 in the presence of single and multiple heavy metals. J. Microbiol. 54, 602–610.
Li, Y., Fang, W., Xie, S., Yang, X., and Wang, L. 2017. Leucobacter corticis sp. nov., isolated from symptomatic bark of Populus × euramericana canker. Int. J. Syst. Evol. Microbiol. 67, 2248–2252.
MIDI. 1999. Sherlock Microbial Identification System Operating Manual, version 3.0. MIDI, Inc., Newark, Delaware, USA.
Morais, P.V., Paulo, C., Francisco, R., Branco, R., Paula Chung, A., and da Costa, M.S. 2006. Leucobacter luti sp. nov., and Leucobacter alluvii sp. nov., two new species of the genus Leucobacter isolated under chromium stress. Syst. Appl. Microbiol. 29, 414–421.
Na, S.I., Kim, Y.O., Yoon, S.H., Ha, S.M., Baek, I., and Chun, J. 2018. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285.
Parte, A.C., Sardà Carbasse, J., Meier-Kolthoff, J.P., Reimer, L.C., and Göker, M. 2020. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612.
Qu, Y., Li, H., Li, A., Ma, F., and Zhou, J. 2011. Identification and characterization of Leucobacter sp. N-4 for Ni (II) biosorption by response surface methodology. J. Hazard. Mater. 190, 869–875.
Ryan, R.P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M.B., Berg, G., van der Lelie, D., and Dow, J.M. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, Delaware, USA.
Schleifer, K.H. and Kandler, O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 36, 407–477.
Schumann, P. and Pukall, R. 2017. Leucobacter weissii sp. nov., an isolate from activated sludge once described as first representative of the peptidoglycan variation B2δ, and emended description of the genus Leucobacter. Int. J. Syst. Evol. Microbiol. 67, 5244–5251.
Shin, N.R., Kim, M.S., Jung, M.J., Roh, S.W., Nam, Y.D., Park, E.J., and Bae, J.W. 2011. Leucobacter celer sp. nov., isolated from Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 61, 2353–2357.
Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Sturm, G., Jacobs, J., Spröer, C., Schumann, P., and Gescher, J. 2011. Leucobacter chromiiresistens sp. nov., a chromate-resistant strain. Int. J. Syst. Evol. Microbiol. 61, 956–960.
Sun, L.N., Pan, D.D., Wu, X.W., Yang, E.D., Hua, R.M., and Li, Q.X. 2018. Leucobacter triazinivorans sp. nov., a s-triazine herbicide prometryn-degrading bacterium isolated from sludge. Int. J. Syst. Evol. Microbiol. 68, 204–210.
Takeuchi, M., Weiss, N., Schumann, P., and Yokota, A. 1996. Leucobacter komagatae gen. nov., sp. nov., a new aerobic Gram-positive, nonsporulating rod with 2,4-diaminobutyric acid in the cell wall. Int. J. Syst. Bacteriol. 46, 967–971.
Teather, R.M. and Wood, P.J. 1982. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43, 777–780.
Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Tindall, B. 1990. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 66, 199–202.
Tittsler, R.P. and Sandholzer, L.A. 1936. The use of semi-solid agar for the detection of bacterial motility. J. Bacteriol. 31, 575–580.
Wadgaonkar, S.L., Nancharaiah, Y.V., Jacob, C., Esposito, G., and Lens, P.N.L. 2019. Microbial transformation of Se oxyanions in cultures of Delftia lacustris grown under aerobic conditions. J. Microbiol. 57, 362–371.
Weon, H.Y., Anandham, R., Tamura, T., Hamada, M., Kim, S.J., Kim, Y.S., Suzuki, K., and Kwon, S.W. 2012. Leucobacter denitrificans sp. nov., isolated from cow dung. J. Microbiol. 50, 161–165.
Xin, H., Itoh, T., Zhou, P., Suzuki, K., Kamekura, M., and Nakase, T. 2000. Natrinema versiforme sp. nov., an extremely halophilic archaeon from Aibi salt lake, Xinjiang, China. Int. J. Syst. Evol. Microbiol. 50, 1297–1303.
Yi, Y.J., Lim, J.M., Gu, S., Lee, W.K., Oh, E., Lee, S.M., and Oh, B.T. 2017. Potential use of lactic acid bacteria Leuconostoc mesenteroides as a probiotic for the removal of Pb(II) toxicity. J. Microbiol. 55, 296–303.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Yun, J.H., Cho, Y.J., Chun, J., Hyun, D.W., and Bae, J.W. 2014. Genome sequence of the chromate-resistant bacterium Leucobacter salsicius type strain M1-8T. Stand. Genomic Sci. 9, 495–504.
Yun, J.H., Roh, S.W., Kim, M.S., Jung, M.J., Park, E.J., Shin, K.S., Nam, Y.D., and Bae, J.W. 2011. Leucobacter salsicius sp. nov., from a salt-fermented food. Int. J. Syst. Evol. Microbiol. 61, 502–506.
Zhu, D., Zhang, P., Li, P., Wu, J., Xie, C., Sun, J., and Niu, L. 2016. Description of Leucobacter holotrichiae sp. nov., isolated from the gut of Holotrichia oblita larvae. Int. J. Syst. Evol. Microbiol. 66, 1857–1861.
Acknowledgments
We thank Dr. Aharon Oren (The Hebrew University of Jerusalem, Israel) and Dr. Bernard Schink (University of Konstanz, Germany) for etymological advice. This work was supported by grants from the Mid-career Researcher Program (NRF-2020R1A2C3012797) funded by the Ministry of Science and ICT, and Basic Science Research Program (NRF-2019R1A6A3A01096031) funded by the Ministry of Education through the National Research Foundation of Korea (NRF). This work was also supported by grants from the National Institute of Biological Resources (NIBR201801106), funded by the Ministry of Environment of Korea (MOE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest The authors declare that there are no conflicts of interest.
Ethical statement All sampling conducted in this study was approved by the Institutional Animal Care and Use Committee of Kyung Hee University (Permit number: KHUASP(SE)-18-048) and complied with guidelines of the Committee.
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hyun, DW., Sung, H., Kim, P.S. et al. Leucobacter coleopterorum sp. nov., Leucobacter insecticola sp. nov., and Leucobacter viscericola sp. nov., isolated from the intestine of the diving beetles, Cybister brevis and Cybister lewisianus, and emended description of the genus Leucobacter. J Microbiol. 59, 360–368 (2021). https://doi.org/10.1007/s12275-021-0472-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-021-0472-6