Abstract
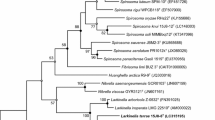
Strain DY6T, a Gram-positive endospore-forming motile rod-shaped bacterium, was isolated from soil in South Korea and characterized to determine its taxonomic position. Phylogenetic analyses based on the 16S rRNA gene sequence of strain DY6T revealed that strain DY6T belongs to the genus Paenibacillus in the family Paenibacillaceae in the class Bacilli. The highest degree of sequence similarities of strain DY6T were found with Paenibacillus gansuensis B518T (97.9%), P. chitinolyticus IFO 15660T (95.3%), P. chinjuensis WN9T (94.7%), and P. rigui WPCB173T (94.7%). Chemotaxonomic data revealed that the predominant fatty acids were anteiso-C15:0 (38.7%) and C16:0 (18.0%). A complex polar lipid profile consisted of major amounts of diphosphatidylglycerol, phosphatidylethanolamine, and phosphatidylglycerol. The predominant respiratory quinone was MK-7. Based on these phylogenetic, chemotaxonomic, and phenotypic data, strain DY6T (=KCTC 33026T =JCM 18491T) should be classified as a type strain of a novel species, for which the name Paenibacillus swuensis sp. nov. is proposed.
Similar content being viewed by others
References
Ash, C., Priest, F.G., and Collins, M.D. 1993. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie van Leeuwenhoek 64, 253–260.
Ash, C., Priest, F.G., and Collins, M.D. 1994. Paenibacillus gen. nov. In Validation of the Publication of New Names and New Combinations Previously Effectively Published Outside the IJSB, List no. 51. Int. J. Syst. Bacteriol. 44, 852.
Baik, K.S., Lim, C.H., Choe, H.N., Kim, E.M., and Seong, C.N. 2011. Paenibacillus rigui sp. nov., isolated from a freshwater wetland. Int. J. Syst. Evol. Microbiol. 61, 529–534.
Collins, M.D. and Jones, D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol. Rev. 45, 316–354.
Ezaki, T., Hashimoto, Y., and Yabuuchi, E. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229.
Felsenstein, J. 1985. Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fitch, W.M. 1971. Toward defining the course of evolution: minimum change for a specified tree topology. Syst. Zool. 20, 406–416.
Gerhardt, P., Murray, R.G.E., Wood, W.A., and Krieg, N.R. 1994. Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, D.C., USA.
Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Im, W.T., Jung, H.M., Ten, L.N., Kim, M.K., Bora, N., Goodfellow, M., Lim, S., Jung, J., and Lee, S.T. 2008. Deinococcus aquaticus sp. nov., isolated from fresh water, and Deinococcus caeini sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 58, 2348–2353.
Kempf, M.J., Chen, F., Kern, R., and Venkateswaran, K. 2005. Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology 5, 391–405.
Kim, O.S., Cho, Y.J., Lee, K., Yoon, S.H., Kim, M., Na, H., Park, S.C., Jeon, Y.S., Lee, J.H., Yi, H., and et al. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721.
Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK.
Komagata, K. and Suzuki, K. 1987. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–207.
Kong, B.H., Liu, Q.F., Liu, M., Liu, Y., Liu, L., Li, C.L., Yu, R., and Li, Y.H. 2013. Paenibacillus typhae sp. nov., isolated from roots of Typha angustifolia L. Int. J. Syst. Evol. Microbiol. 63, 1037–1044.
Lanyi, B. 1987. Classical and rapid identification methods for medically important bacteria. Methods Microbiol. 19, 1–67.
Lee, L.J., Lee, H.J., Jang, G.S., Yu, J.M., Cha, J.Y., Kim, S.J., Lee, E.B., and Kim, M.K. 2013. Deinococcus swuensis sp. nov., a gamma-radiation-resistant bacterium isolated from soil. J. Microbiol. 51, 305–311.
Lee, J.S., Pyun, Y.R., and Bae, K.S. 2004. Transfer of Bacillus ehimensis and Bacillus chitinolyticus to the genus Paenibacillus with emended descriptions of Paenibacillus ehimensis comb. nov. and Paenibacillus chitinolyticus comb. nov. Int. J. Syst. Evol. Microbiol. 54, 929–933.
Lim, J.M., Jeon, C.O., Lee, J.C., Xu, L.H., Jiang, C.L., and Kim, C.J. 2006a. Paenibacillus gansuensis sp. nov., isolated from desert soil of Gansu Province in China. Int. J. Syst. Evol. Microbiol. 56, 2131–2134.
Lim, S., Yoon, H., Ryu, S., Jung, J., Lee, M., and Kim, D. 2006b. A comparative evaluation of radiation-induced DNA damage using real-time PCR: influence of base composition. Radiat. Res. 165, 430–437.
Lim, S., Song, D., Joe, M., and Kim, D. 2012. Development of a qualitative dose indicator for gamma radiation using lyophilized Deinococcus. J. Microbiol. Biotechnol. 22, 1296–1300.
Mesbah, M., Premachandran, U., and Whitman, W.B. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39, 159–167.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, DE, USA.
Shin, Y.K., Lee, J.S., Chun, C.O., Kim, H.J., and Park, Y.H. 1996. Isoprenoid quinone profiles of the Leclercia adecarboxylata KCTC 1036T. J. Microbiol. Biotechnol. 6, 68–69.
Smibert, R.M. and Krieg, N.R. 1994. Phenotypic characterization. Methods for General and Molecular Bacteriology, pp. 607–654. In Gerhardt, P. (ed.). American Society for Microbiology, Washington, D.C., USA.
Srinivasan, S., Kim, M.K., Lim, S., Joe, M., and Lee, M. 2012a. Deinococcus daejeonensis sp. nov., isolated from sludge in a sewage disposal plant. Int. J. Syst. Evol. Microbiol. 62, 1265–1270.
Srinivasan, S., Lee, J.J., Lim, S., Joe, M., and Kim, M.K. 2012b. Deinococcus humi sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 62, 2844–2850.
Stackebrandt, E. and Goebel, B.M. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849.
Tamaoka, J. and Komagata, K. 1984. Determination of DNA base composition by reversed phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882.
Wayne, L.G., Brenner, D.J., Colwell, R.R., Grimont, P.A.D., Kandler, O., Krichevsky, M.I., Moore, L.H., Moore, W.E.C., Murray, R.G.E., Stackebrandt, E., and et al. 1987. International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane, D.J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Yoon, J.H., Seo, W.T., Shin, Y.K., Kho, Y.H., Kang, K.H., and Park, Y.H. 2002. Paenibacillus chinjuensis sp. nov., a novel exopolysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 52, 415–421.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
The NCBI GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain DY6T (=KCTC 33026T =JCM 18491T) is JQ958374.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lee, JJ., Yang, DH., Ko, YS. et al. Paenibacillus swuensis sp. nov., a bacterium isolated from soil. J Microbiol. 52, 106–110 (2014). https://doi.org/10.1007/s12275-014-3546-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-014-3546-x