Abstract
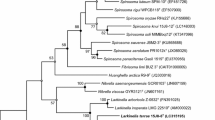
Strain 18JY21-1T, a Gram-positive, endospore-forming, motile, and rod-shaped bacterium, was isolated from soil in South Korea and was characterised to determine its taxonomic position. Phylogenetic analysis based on the 16S rRNA gene sequence of strain 18JY21-1T revealed that the strain 18JY21-1T belongs to the genus Paenibacillus in the family Paenibacillaceae in the class Bacilli. The highest degree of sequence similarities of strain 18JY21-1T was found with Paenibacillus doosanensis CAU 1055T (97.7%) and Paenibacillus protaetiae KACC 19327T (94.4%). In genome analysis, the calculated average nucleotide identity (ANI) and the digital DNA–DNA hybridization (DDH) values between strain 18JY21-1T and Paenibacillus protaetiae KACC 19327T were 66.3% and 22.8%, respectively. Chemotaxonomic data revealed that the predominant fatty acids were anteiso-C15:0 (38.7%) and C16:0 (18.0%). A complex polar lipid profile consisted of major amounts of diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG). The cell wall peptidoglycan was meso-diaminopimelic acid. The predominant respiratory quinone was MK-7. Based on the phylogenetic, chemotaxonomic, and phenotypic data, strain 18JY21-1T (= KCTC 3396T = JCM 33183T) should be classified as a type strain of a novel species, for which the name Paenibacillus albiflavus sp. nov. is proposed.


Similar content being viewed by others
References
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie Van Leeuwenhoek 64:253–260
Ash C, Priest FG, Collins MD (1994) Paenibacillus gen. nov. In Validation of the Publication of New Names and New Combinations Previously Effectively Published Outside the IJSB, List no. 51. Int J Syst Bacteriol 44:852. https://doi.org/10.1099/00207713-44-4-852
Baik KS, Lim CH, Choe HN, Kim EM, Seong CN (2011) Paenibacillus rigui sp. nov., isolated from a freshwater wetland. Int J Syst Evol Microbiol 61:529–534
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Chung YR, Kim CH, Hwang I, Chun J (2000) Paenibacillus koreensis sp. nov., a new species that produces an iturin-like antifungal compound. Int J Syst Evol Microbiol 50:1495–1500
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev 45:316–354
Devereux R, He SH, Doyle CL, Orkland S, Stahl DA, LeGall J, Whitman WB (1990) Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J Bacteriol 172:3609–3619
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20(4):406–416
Gerhardt P, Murray RGE, Wood WA, Krieg NR (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington
Grady EN, MacDonald J, Liu L et al (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact 15:203
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Im WT, Jung HM, Ten LN, Kim MK, Bora N et al (2008) Deinococcus aquaticus sp. nov., isolated from fresh water, and Deinococcus caeni sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 58:2348–2353
Kempf MJ, Chen F, Kern R, Venkateswaran K (2005) Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology 5:391–405
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol Mar 62(Pt3):716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kong BH, Liu QF, Liu M, Liu Y, Liu L et al (2013) Paenibacillus typhae sp. nov., isolated from roots of Typha angustifolia L. Int J Syst Evol Microbiol 63:1037–1044
Lányi B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Lee I, Kim YO, Park SC, Chun J (2015) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Priest FG (1977) Extracellular enzyme synthesis in the genus Bacillus. Bacteriol Rev 41:711–753
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106(45):19126–19131
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI Inc, Newark
Seo WT, Kahng GG, Nam SH, Choi SD, Suh HH et al (1999) Isolation and characterization of a novel exopolysaccharide-producing Paenibacillus sp. WN9 KCTC 8951P. J Microbiol Biotechnol 9:820–825
Shin YK, Lee JS, Chun CO, Kim HJ, Park YH (1996) Isoprenoid quinone profiles of the Leclercia adecarboxylata KCTC 1036T. J Microbiol Biotechnol 6:68–69
Slepecky RA, Hemphill HE (1991) The genus Bacillus nonmedical. In: Balows A, Triiper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes. Springer, New York
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P (ed) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Acknowledgements
This research was supported by the research grant from Seoul Women’s University (2021), and MISP (Ministry of Science, ICT & Future Planning), Korea, under the National Program for Excellence in SW supervised by the IITP (Institute of Information & communications Technology Planning & Evaluation) (2016-0-00022) and by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR201801204)
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, J.H., Lee, S.E. & Srinivasan, S. Paenibacillus albiflavus sp. nov., a bacterium isolated from soil. Arch Microbiol 203, 4973–4979 (2021). https://doi.org/10.1007/s00203-021-02476-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02476-3