Abstract
The apolipoprotein E (APOE) genotype is an important risk factor for ageing and age-related diseases. The APOE4 genotype (in contrast to APOE3) has been shown to be associated with oxidative stress and chronic inflammation. Metallothioneins (MT) exhibit antioxidant and anti-inflammatory activity, and MT overexpression has been shown to increase lifespan in mice. Interactions between APOE and MT, however, are largely unknown. Hence, we determined the effect of the APOE4 versus APOE3 genotype on MT levels in targeted gene replacement mice. APOE4 versus APOE3 mice exhibited significantly lower hepatic MT1 and MT2 mRNA as well as lower MT protein levels. The decrease in hepatic MT protein levels in APOE4 as compared to APOE3 mice was accompanied by lower nuclear Nrf1, a protein partly controlling MT gene expression. Cell culture experiments using hepatocytes identified allyl-isothiocyanate (AITC) as a potent MT inductor in vitro. Therefore, we supplemented APOE3 and APOE4 mice with AITC. However, AITC (15 mg/kg b.w.) could only partly correct for decreased MT1 and MT2 gene expression in APOE4 mice in vivo. Furthermore, cholesterol significantly decreased both Nrf1 and MT mRNA levels in Huh7 cells indicating that differences in MT gene expression between the two genotypes could be related to differences in hepatic cholesterol concentrations. Overall, present data suggest that the APOE genotype is an important determinant of tissue MT levels in mice and that MT gene expression may be impaired by the APOE4 genotype.
Similar content being viewed by others
Introduction
Apolipoprotein E (APOE) is a 34-kDa protein, containing 299 amino acids, which is mainly synthesized by the liver but also in brain and macrophages (Newman et al. 1985; Williams et al. 1985; Basu et al. 1981). The APOE gene is polymorphic with three common alleles, coding for three major APOE isoforms designated APOE2, APOE3 and APOE4. The APOE isoforms differ in the amino acids at positions 112 and 158 of the mature protein (Weisgraber et al. 1981). APOE2 has two cysteines, APOE3 has Cys-112 and Arg-158, and APOE4 has two arginines in these positions. The three alleles differ in their frequencies: APOE3 represents 65–70%, while APOE4 carriers account for up to 15–20% of the Caucasian population (Mahley et al. 2009). Presence of the APOE4 allele is associated with a significantly higher risk of age-related chronic diseases including coronary heart disease (CHD) (Humphries et al. 2001; Stephens et al. 2008) and Alzheimer’s disease (AD) (Corder et al. 1993). Cohort studies revealed that the E4 allele of APOE was significantly less frequent in centenarians than in younger controls (Schachter et al. 1994). There is increasing evidence demonstrating that APOE4 may be associated with elevated oxidative stress (Dietrich et al. 2005) and chronic inflammation (Jofre-Monseny et al. 2007), which may in turn contribute to the increased CHD and AD risk in this subgroup of the population.
Metallothioneins (MT) are low molecular weight cysteine-rich intracellular proteins with a high binding affinity for essential transition metals, such as zinc and copper. Metallothioneins exist in different isoforms characterized by the length of amino acid chain (MT1–MT4), but MT1 and MT2 are the most widely distributed MT isoforms (Swindell 2011). MT1 and MT2 are expressed in numerous tissues including liver, kidney, heart and lung. MT gene transcription is rapidly induced by transition and heavy metals as well as by oxidative stress (Dalton et al. 1994; Palmiter 1987). MT exhibit antioxidant properties (Miles et al. 2000) and may play a role in the prevention of atherosclerosis (Giacconi et al. 2008). MT might prevent high fat diet-induced cardiac dysfunction (Dong et al. 2007), and overexpression of MT in the heart has been demonstrated to increase lifespan in mice (Yang et al. 2006). Furthermore, overexpression of metal responsive transcription factor 1 (Mtf1) ameliorated lifespan reductions associated with oxidative stress in Drosophila melanogaster (Bahadorani et al. 2010). Nevertheless, the functional role of MT in the ageing process still remains to be elucidated (Mocchegiani et al. 2011).
The MT promoter comprises several response elements that regulate its transcription, including the metal response elements (MRE), which are activated by Mtf1 in the presence of dietary zinc and copper (Radtke et al. 1993; Bittel et al. 1998). Furthermore, the antioxidant response element (ARE) mediates MT expression in response to reactive oxygen species and may act synergistically with MRE (Dalton et al. 1994; Andrews 2000). The nuclear factors erythroid-derived 2-related factor 1 (Nrf1) and factor 2 (Nrf2), both members of the Cap ‘n’ collar (CNC) subfamily of basic leucine zipper (ZIP) transcription factors, bind to ARE (Biswas and Chan 2010; Venugopal and Jaiswal 1996; Brigelius-Flohe and Flohe 2011). Recent data indicate that the expression of MT1 and MT2 genes is largely dependent on Nrf1 (Ohtsuji et al. 2008). Both Nrf1 and Nrf2 bind to ARE, but only Nrf1 may form a complex that activates transcription of the MT1 gene. Little is known about the interaction between APOE and MT, both proteins centrally involved in the ageing process. Therefore, we investigated the effect of the APOE genotype on MT levels in APOE3 and APOE4 targeted gene replacement mice.
Allyl-isothiocyanate (AITC) belongs to the family of isothiocyanates and occurs in many cruciferous vegetables such as cauliflower, Brussels sprouts, cabbage and kale (Kushad et al. 1999; Rungapamestry et al. 2006). AITC is derived from its precursor sinigrin (a glucosinolate) following myrosinase-catalysed hydrolysis (Krul et al. 2002; Bhattacharya et al. 2010). AITC directly interacts with sulfhydryl groups (Zhang et al. 2005) and predominantly conjugates with cysteine residues (Zhang et al. 2010). AITC has been shown to induce several phase II and antioxidant enzymes, including GST, HO-1 and NQO1 (Zhang and Talalay 1998; Ye and Zhang 2001; Tang and Zhang 2004; Jeong et al. 2005; Bogaards et al. 1990; McWalter et al. 2004). Since AITC has been suggested to be less cytotoxic than sulforaphane (Wagner et al. 2011), another isothiocyanate that has been recently reported to induce MT in human hepatocytes (Hu et al. 2004; Yeh and Yen 2005), we investigated whether dietary AITC may induce MT levels in Huh7 cells and could correct for decreased hepatic MT and Nrf1 mRNA levels in APOE4 mice.
Materials and methods
Mice and diets
Female homozygous APOE3 and APOE4 targeted replacement mice (6–8 weeks old) ‘humanized’ for the APOE gene were purchased from Taconic Europe (Ry, Denmark). In order to verify respective APOE genotypes of the humanized targeted replacement mice, the two single-nucleotide polymorphisms in the APOE gene (rs429358, rs7412) were determined using TaqMan® SNP genotyping assays by Applied Biosystems according to manufacturer’s instructions (Carlsbad CA, USA). In brief, 40× TaqMan® SNP genotyping assay was diluted 1:2 in TE buffer. Subsequently, the 20× TaqMan® SNP genotyping assay was mixed with 2× TaqMan® Universal Mastermix, bidest. water and DNA to a 1× dilution and a final reaction volume of 10 μl. For each reaction, 20 ng DNA was implemented. Control samples with known APOE genotypes were included. PCR was performed using a Rotor Gene 3000 thermal cycler (Corbett research). Liver APOE levels of the two isoproteins were not different between the two genotypes. Mice were kept in macrolon cages (3–4 animals per cage) at 21–25°C with a 12-h day–night cycle and according to the German Regulations for Animal Welfare approved by the local authority (MLUR, Kiel, Germany, No. V312-72241.121-33 (88-7/09)). Semi-synthetic diets (‘Western-type diets’), based on corn starch (14.5%), casein (17.1%), sucrose (32.8%) and butter fat (21.2%), containing 48 mg iron, 39 mg zinc and 11 mg copper per kg diet were purchased from Ssniff Special Diets (Soest, Germany). Diets and water were provided ad libitum. Study 1 was conducted using eight APOE3 and eight APOE4 mice per group, respectively. Study 1 lasted 6 weeks. Samples of study 1 were used for MT, Nrf1, Mtf1 as well as zinc, copper and iron analysis in response to the APOE genotype. Study 2 was carried out with a two-factorial experimental design. One group of each APOE genotype was supplemented with either 15 mg AITC/kg body weight (Sigma-Aldrich, Steinheim, Germany) or solvent (PBS) by oral gavage for 7 days. Study 2 was conducted to determine whether dietary AITC may correct for decreased hepatic MT and Nrf1 mRNA levels in APOE4 mice. The experimental designs of mouse study 1 and 2 are summarized in Table 1.
At the end of the experimental trial, mice were fasted for 12 h prior anaesthesia with carbon dioxide and killed by decapitation. In both mouse studies, food intake (recorded daily) and final body weights (recorded weekly) were not significantly different between groups.
Tissue preparation
Liver, heart, kidney and lung were excised and dissected, one part was stored in RNAlater™ (Qiagen, Hilden, Germany), and the remainder was snap-frozen in liquid nitrogen and stored at −80°C prior usage.
Cell culture
Human hepatoma cells (Huh7; Institute of Applied Cell Culture, Munich, Germany) were maintained in Dulbecco’s modified Eagle’s medium containing 4.5 g/l glucose, 4 mmol/l l-glutamine, 1 mmol/l sodium pyruvate, 10% foetal calf serum (FCS), 100 U/ml penicillin and 100 μg/ml streptomycin (all PAA, Coelbe, Germany). Cells were grown in 5% CO2 at 37°C under a humidified atmosphere. AITC was dissolved in DMSO, and cholesterol was dissolved in ethanol. Final solvent concentrations in cell culture medium were <0.1%.
RNA isolation and real-time PCR
Huh7 cells were seeded at an initial density of 250,000 cells/well in 12-well plates and incubated with medium supplemented with either AITC (10, 25 μmol/l) or 20 mg/ml cholesterol for 6 h. RNA from Huh7 cells as well as from liver, heart, kidney and lung of APOE3 and APOE4 transgenic mice (20–30 mg) was isolated with TRIsure following the manufacturer’s instructions (Bioline, Luckenwalde, Germany) and quantified photometrically (Spectrophotometer DU800; Beckman Coulter, Krefeld, Germany). Primers for murine and human MT1, MT2, Nrf1, GAPDH and β-actin (Table 2) were designed by standard tools (Spidey, Primer3, NCBI Blast) and purchased from MWG (Ebersberg, Germany). Quantitative real-time PCR was performed as one-step procedure using Sensi-Mix™ one-step kit (Quantace, Berlin, Germany) with SybrGreen detection using the Rotorgene 6000 cycler (Corbett Life Science, Sydney, Australia). Quantitation was done by the use of a standard curve. Transcription levels of target genes were related to transcription of the housekeeping gene GAPDH or β-actin.
Western blot analysis
Liver nuclear extracts for Western blotting were prepared as described in Wagner et al. (Wagner et al. 2010). Membranes were probed with antibodies against Nrf1 and Mtf1 (1:500; Santa Cruz Biotechnology, Heidelberg, Germany). Target band densities were normalized to the loading control TATA-binding protein (TBP) (1:500; Santa Cruz Biotechnology, Heidelberg, Germany).
109Cd affinity assay
Total MT was estimated indirectly with the 109Cd-haemoglobin binding assay and gamma counting as described previously (Eaton and Toal 1982).
ICP-AES
Zinc, copper and iron concentrations in liver were determined by inductively coupled plasma atomic emission spectrometry (ICP-AES, Unicam 701) as described previously (Rimbach and Pallauf 1997).
Promoter DNA methylation analysis
The presence of CpG islands within the MT1 and MT2 gene was predicted using the EMBL-EBI Open Software Suite CpGplot (http://www.ebi.ac.uk/Tools/emboss/cpgplot/). Quantitative methylation analysis of the two genes was performed with the MassARRAY® system (Sequenom, Hamburg, Germany) at Bioglobe (Hamburg, Germany) as described previously (Fischer et al. 2010). The murine H19 gene region served as control (data not shown).
Statistical analysis
The statistical analysis was performed with SPSS Version 15.0 (SPSS GmbH Software, Munich, Germany). For the comparison of two groups, tissue (Study 1) and cell culture data were tested for normal distribution (Kolmogorow-Smirnov and Shapiro-Wilk test) and analysed by Student’s t-test. For the comparison of cell culture and mice data (Study 2) with more than two experimental groups, one-way ANOVA was performed following post hoc tests (Dunnett-T or Games-Howell). In the case of non-parametric data, Mann-Whitney U-test was performed. Results are expressed as means with standard errors (SEM), and significance was accepted at p < 0.05.
Results
In study 1, we determined the effect of the APOE genotype on tissue metallothionein, Nrf1 and Mtf1 levels as well as hepatic zinc, copper and iron concentrations in APOE3 and APOE4 targeted gene replacement mice.
Effect of APOE genotype on hepatic metallothionein
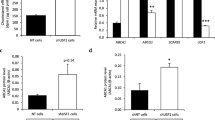
We found significantly lower mRNA steady-state levels of MT1 and MT2 in the liver of APOE4 in comparison with APOE3 mice (Fig. 1a, b). The hepatic MT protein level as measured by the 109Cd affinity assay was significantly decreased in APOE4 versus APOE3 mice (Fig. 1c). Interestingly, MT1 and MT2 exhibit CpG islands in their promoter regions, suggesting putative regulatory functions through methylation in these areas. Since we found lower hepatic mRNA levels of MT1 and MT2 in APOE4 compared to APOE3 mice, we quantified the methylation status of MT1 and MT2 CpG islands in the liver of APOE4 versus APOE3 mice. The resulting epigrams revealed that all samples exhibited similar methylation patterns. We found no quantitative differences in the promoter methylation of the MT1 and MT2 gene between APOE4 and APOE3 mice (Fig. 1d).
Effect of APOE genotype on hepatic metallothionein 1 (MT1; a) and 2 (MT2; b) mRNA levels (n = 8) determined by real-time PCR as well as metallothionein protein level (n = 8) (c) as measured by 109Cd affinity assay in APOE3 (open bars) as compared to APOE4 mice (solid bars). Data are means + SEM (study 1). Asterisks (*) indicate significant differences between APOE3 and APOE4 animals. Genomic DNA isolated from the liver of APOE3 and APOE4 mice was analysed for methylation status of cytosine-phospho-guanine (CpG) sites in the metallothionein 1 (MT1) and 2 (MT2) gene promoters (d). A 255-bp portion of the H19 regulatory region served as control amplicon. Exemplary data for one amplicon for each gene are shown. The grey dots indicate the software-determined methylation ratio at each analysed CpG unit for each sample. Upper numbering scheme refers to the base positions within the analysed amplicon. Lower numbers indicate the individual CpG sites located in the amplicon
Effect of APOE genotype on hepatic Nrf1 and Mtf1
Since gene expression of MT1 and MT2 is, at least partly, controlled by the transcription factors Nrf1 and Mtf1, we determined both mRNA and nuclear protein levels of Nrf1 and Mtf1 in the liver of both APOE genotypes. We found significantly lower Nrf1 mRNA levels (Fig. 2a) and lower nuclear Nrf1 protein levels in the liver of APOE4 versus APOE3 mice (Fig. 2b). However, no significant differences in Mtf1 mRNA and nuclear protein levels of Mtf1 were found between APOE4 and APOE3 mice (data not shown).
Effect of APOE genotype on hepatic Nrf1 mRNA (a) and nuclear protein (b) level in APOE3 (open bars) as compared to APOE4 mice (solid bars). Data of mRNA levels as determined by real-time PCR are means + SEM, n = 8 per group (study 1). Asterisk (*) indicates significant differences between APOE3 and APOE4 mice; effect of APOE genotype on hepatic zinc (c), copper (d) and iron (e) concentrations as determined by ICP-AES. Data are means + SEM, n = 8 per group (study 1)
Hepatic zinc, copper and iron concentrations
Since transition metals, including zinc and copper, may induce metallothionein expression, we determined zinc and copper levels in the liver of our mice in response to the APOE genotype. Furthermore, zinc and copper may affect liver iron levels. However, under our study conditions, neither zinc nor copper or iron concentrations in the liver were significantly affected by the APOE genotype (Fig. 2c–e).
Effect of APOE genotype on metallothionein and Nrf1 levels in heart, kidney and lung
As differences in hepatic gene expression between APOE4 and APOE3 mice were evident, we determined MT1, MT2 and Nrf1 mRNA levels also in heart, kidney and lung in response to the APOE genotype. In contrast to APOE3 mice, MT1, MT2 and Nrf1 mRNA levels were significantly lower in heart (Fig. 3a–c), kidney (Fig. 3d–f) and lung (Fig. 3g–i) of APOE4 animals.
Effect of APOE genotype on metallothionein 1 (MT1) and 2 (MT2) and Nrf1 mRNA levels (n = 8) as determined by real-time PCR in the heart (a–c), kidney (d–f) and lung (g–i) of APOE3 (open bars) as compared to APOE4 mice (solid bars). Data are means + SEM. Asterisks (*) indicate significant differences between APOE3 and APOE4 mice (study 1)
Effect of AITC on MT and Nrf1 mRNA level in Huh7 cells and liver of APOE transgenic mice
Isothiocyanates are potent inducers of metallothioneins both in cultured cells (Ernst et al. 2011) and in laboratory rodents (Hu et al. 2004). As shown in Fig. 4a, allyl-isothiocyanate (AITC) treatment of Huh7 cells at non-cytotoxic concentrations resulted in a dose-dependent increase in MT2 levels. Based on our cell culture data, we performed an additional mouse study (study 2) investigating whether dietary AITC may correct for decreased hepatic MT1, MT2 and Nrf1 mRNA levels in APOE4 mice. In agreement with mouse study 1, APOE4 mice exhibited lower hepatic MT1, MT2 and Nrf1 mRNA levels as compared to APOE3 mice in study 2 (Fig. 4b–d). Dietary AITC could only partly counteract decreased Nrf1 and MT2 but not MT1 gene expression in our APOE4 mice.
Effect of 10 and 25 μmol/l AITC on MT2 (a) mRNA level in Huh7 cells. Data are means + SEM, n = 6. Asterisk (*) indicates significant differences between untreated controls and AITC-treated cells; Effect of AITC and APOE genotype on hepatic metallothionein 1 (MT1; b) and metallothionein 2 (MT2; c) and Nrf1 (d) mRNA levels determined by real-time PCR in APOE3 (open bars) as compared to APOE4 mice (solid bars) fed with and without 15 mg AITC/kg body weight. Values are means + SEM, n = 6 (study 2). Means without a common letter significantly differ, p < 0.05
Effect of cholesterol on metallothionein and Nrf1 levels in Huh7 cells
Treatment of Huh7 cells with cholesterol (at a non-cytotoxic concentration) significantly decreased MT2 as well as Nrf1 levels in Huh7 hepatocytes as summarized in Fig. 5.
Discussion
An important finding of our study is that the APOE4 genotype, in contrast to APOE3, is associated with decreased MT levels in mice liver, heart, kidney and lung. MT exhibits antioxidant (Miles et al. 2000) and anti-inflammatory properties (Inoue et al. 2009). We have previously shown that the APOE4 genotype is associated with an altered inflammatory response (Jofre-Monseny et al. 2007) and differences in the cellular oxidant/antioxidant status (Dietrich et al. 2005; Jofre-Monseny et al. 2008a; Huebbe et al. 2007) in cultured cells, mice and humans. These differences in the inflammatory and oxidant/antioxidant status between the two genotypes may be partly related to differences in metallothionein levels.
The underlying cellular and molecular mechanisms by which APOE isoforms may affect tissue metallothionein levels are largely unknown. Our promoter DNA methylation analyses suggest that differences in MT gene expression between APOE3 and APOE4 mice are most likely not related to differences in the methylation status of both the MT1 and MT2 promoter. Furthermore, we did not observe differences in hepatic zinc and copper concentrations, both known inducers of MT, between the two genotypes. In this study, total liver zinc was determined by ICP-AES. However, we did not measure the free zinc pool in our mice. A recent study suggests that APOE may affect the free zinc pool since the level of histochemically reactive zinc was reduced in the brain of APOE-deficient mice compared to wild-type animals (Lee et al. 2010). Thus, future studies are warranted to test the hypothesis whether the free zinc pool may be differentially regulated in response to the APOE genotype.
We observed lower nuclear Nrf1 levels in APOE4 versus APOE3 mice. Nrf1 is an important molecular switch regulating tissue MT concentration (Ohtsuji et al. 2008). The decreased Nrf1 and MT levels in APOE4 versus APOE3 may be due to differences in hepatic cholesterol concentrations between APOE3 and APOE4 mice. We have previously shown that APOE4 as compared to APOE3 mice exhibit elevated liver cholesterol levels (Graeser et al. 2011). Higher hepatic cholesterol levels in APOE4 mice may be due to a defective cholesterol reuptake into the hepatic tissue (Carvalho-Wells et al. 2010). Therefore, we supplemented Huh7 cells with cholesterol and determined Nrf1 and MT2 levels, which are highly expressed in this cell line. Interestingly, cholesterol significantly decreased both Nrf1 and MT2 mRNA levels (Fig. 5), indicating that differences in MT gene expression between the two genotypes could be partly related to differences in hepatic cholesterol concentrations.
Since APOE4 versus APOE3 mice exhibited lower tissue MT levels, we asked the question whether this could be corrected by dietary factors. Yeh and Yen (Yeh and Yen 2005) reported that the isothiocyanate sulforaphane (SFN) induces MT in HepG2 hepatocytes. Our recent cell culture data suggest that allyl-isothiocyanate (AITC) is less cytotoxic than SFN but almost equally potent than SFN as far its anti-inflammatory properties and the induction of stress response genes are concerned (Wagner et al. 2011). Thus, we tested AITC for its ability to induce MT2 in Huh7 hepatocytes. Similar to the effects described for SFN (Yeh and Yen 2005), we observed a dose-dependent induction of MT2 by AITC in the present cell culture study. However, in our mouse study, AITC could only partly correct for decreased gene expression in APOE4 versus APOE3 mice. In the present study, mice were supplemented with AITC only for 1 week and only one AITC concentration was chosen. It needs to be established if long-term administration and higher doses of AITC may be more effective for inducing MT gene expression in APOE4 mice. Overall, APOE4 mice appear to be not very responsive towards Nrf1 and MT induction by AITC. This is in accordance with our previous findings in mice using flavonoids, indicating that APOE4 versus APOE3 mice exhibited lower paraoxonase-1 levels that could be better induced by dietary quercetin in APOE3 as compared to APOE4 mice (Boesch-Saadatmandi et al. 2010).
APOE4 affects both morbidity and mortality in humans (Mahley et al. 2009; Jofre-Monseny et al. 2008b). A dysregulation in MT homoeodynamics may adversely affect age-related chronic diseases (Swindell 2011). Here, we showed for the first time, a significant interaction between APOE genotype and MT levels in mice. Future studies are needed to investigate how this APOE/MT interaction affects stress response, chronic disease risk and longevity.
References
Andrews GK (2000) Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol 59(1):95–104
Bahadorani S, Mukai S, Egli D, Hilliker AJ (2010) Overexpression of metal-responsive transcription factor (MTF-1) in Drosophila melanogaster ameliorates life-span reductions associated with oxidative stress and metal toxicity. Neurobiol Aging 31(7):1215–1226
Basu SK, Brown MS, Ho YK, Havel RJ, Goldstein JL (1981) Mouse macrophages synthesize and secrete a protein resembling apolipoprotein E. Proc Natl Acad Sci USA 78(12):7545–7549
Bhattacharya A, Li Y, Wade KL, Paonessa JD, Fahey JW, Zhang Y (2010) Allyl isothiocyanate-rich mustard seed powder inhibits bladder cancer growth and muscle invasion. Carcinogenesis 31(12):2105–2110
Biswas M, Chan JY (2010) Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol 244(1):16–20
Bittel D, Dalton T, Samson SL, Gedamu L, Andrews GK (1998) The DNA binding activity of metal response element-binding transcription factor-1 is activated in vivo and in vitro by zinc, but not by other transition metals. J Biol Chem 273(12):7127–7133
Boesch-Saadatmandi C, Niering J, Minihane AM, Wiswedel I, Gardeman A, Wolffram S, Rimbach G (2010) Impact of apolipoprotein E genotype and dietary quercetin on paraoxonase 1 status in apoE3 and apoE4 transgenic mice. Atherosclerosis 211(1):110–113
Bogaards JJ, van Ommen B, Falke HE, Willems MI, van Bladeren PJ (1990) Glutathione S-transferase subunit induction patterns of Brussels sprouts, allyl isothiocyanate and goitrin in rat liver and small intestinal mucosa: a new approach for the identification of inducing xenobiotics. Food Chem Toxicol 28(2):81–88
Brigelius-Flohe R, Flohe L (2011) Basic principles and emerging concepts in the redox control of transcription factors. Antioxid Redox Signal 15(8):2335–2381
Carvalho-Wells AL, Jackson KG, Gill R, Olano-Martin E, Lovegrove JA, Williams CM, Minihane AM (2010) Interactions between age and apoE genotype on fasting and postprandial triglycerides levels. Atherosclerosis 212(2):481–487
Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261(5123):921–923
Dalton T, Palmiter RD, Andrews GK (1994) Transcriptional induction of the mouse metallothionein-I gene in hydrogen peroxide-treated Hepa cells involves a composite major late transcription factor/antioxidant response element and metal response promoter elements. Nucleic Acids Res 22(23):5016–5023
Dietrich M, Hu Y, Block G, Olano E, Packer L, Morrow JD, Hudes M, Abdukeyum G, Rimbach G, Minihane AM (2005) Associations between apolipoprotein E genotype and circulating F2-isoprostane levels in humans. Lipids 40(4):329–334
Dong F, Li Q, Sreejayan N, Nunn JM, Ren J (2007) Metallothionein prevents high-fat diet induced cardiac contractile dysfunction: role of peroxisome proliferator activated receptor gamma coactivator 1alpha and mitochondrial biogenesis. Diabetes 56(9):2201–2212
Eaton DL, Toal BF (1982) Evaluation of the Cd/hemoglobin affinity assay for the rapid determination of metallothionein in biological tissues. Toxicol Appl Pharmacol 66(1):134–142
Ernst IM, Wagner AE, Schuemann C, Storm N, Hoppner W, Doring F, Stocker A, Rimbach G (2011) Allyl-, butyl- and phenylethyl-isothiocyanate activate Nrf2 in cultured fibroblasts. Pharmacol Res 63(3):233–240
Fischer A, Gaedicke S, Frank J, Doring F, Rimbach G (2010) Dietary vitamin E deficiency does not affect global and specific DNA methylation patterns in rat liver. Br J Nutr 104(7):935–940
Giacconi R, Caruso C, Malavolta M, Lio D, Balistreri CR, Scola L, Candore G, Muti E, Mocchegiani E (2008) Pro-inflammatory genetic background and zinc status in old atherosclerotic subjects. Ageing Res Rev 7(4):306–318
Graeser AC, Boesch-Saadatmandi C, Lippmann J, Wagner AE, Huebbe P, Storm N, Hoppner W, Wiswedel I, Gardemann A, Minihane AM, Doring F, Rimbach G (2011) Nrf2-dependent gene expression is affected by the proatherogenic apoE4 genotype-studies in targeted gene replacement mice. J Mol Med 89(10):1027–1035
Hu R, Hebbar V, Kim BR, Chen C, Winnik B, Buckley B, Soteropoulos P, Tolias P, Hart RP, Kong AN (2004) In vivo pharmacokinetics and regulation of gene expression profiles by isothiocyanate sulforaphane in the rat. J Pharmacol Exp Ther 310(1):263–271
Huebbe P, Jofre-Monseny L, Boesch-Saadatmandi C, Minihane AM, Rimbach G (2007) Effect of apoE genotype and vitamin E on biomarkers of oxidative stress in cultured neuronal cells and the brain of targeted replacement mice. J Physiol Pharmacol 58(4):683–698
Humphries SE, Talmud PJ, Hawe E, Bolla M, Day IN, Miller GJ (2001) Apolipoprotein E4 and coronary heart disease in middle-aged men who smoke: a prospective study. Lancet 358(9276):115–119
Inoue K, Takano H, Shimada A, Satoh M (2009) Metallothionein as an anti-inflammatory mediator. Med Inflamm 2009:101659
Jeong WS, Keum YS, Chen C, Jain MR, Shen G, Kim JH, Li W, Kong AN (2005) Differential expression and stability of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural chemopreventive compounds in HepG2 human hepatoma cells. J Biochem Mol Biol 38(2):167–176
Jofre-Monseny L, Loboda A, Wagner AE, Huebbe P, Boesch-Saadatmandi C, Jozkowicz A, Minihane AM, Dulak J, Rimbach G (2007) Effects of apoE genotype on macrophage inflammation and heme oxygenase-1 expression. Biochem Biophys Res Commun 357(1):319–324
Jofre-Monseny L, Huebbe P, Stange I, Boesch-Saadatmandi C, Frank J, Jackson K, Minihane AM, Rimbach G (2008a) Influence of apolipoprotein E genotype and dietary alpha-tocopherol on redox status and C-reactive protein levels in apolipoprotein E3 and E4 targeted replacement mice. Br J Nutr 100(1):44–53
Jofre-Monseny L, Minihane AM, Rimbach G (2008b) Impact of apoE genotype on oxidative stress, inflammation and disease risk. Mol Nutr Food Res 52(1):131–145
Krul C, Humblot C, Philippe C, Vermeulen M, van Nuenen M, Havenaar R, Rabot S (2002) Metabolism of sinigrin (2-propenyl glucosinolate) by the human colonic microflora in a dynamic in vitro large-intestinal model. Carcinogenesis 23(6):1009–1016
Kushad MM, Brown AF, Kurilich AC, Juvik JA, Klein BP, Wallig MA, Jeffery EH (1999) Variation of glucosinolates in vegetable crops of Brassica oleracea. J Agric Food Chem 47(4):1541–1548
Lee JY, Cho E, Kim TY, Kim DK, Palmiter RD, Volitakis I, Kim JS, Bush AI, Koh JY (2010) Apolipoprotein E ablation decreases synaptic vesicular zinc in the brain. Biometals 23(6):1085–1095
Mahley RW, Weisgraber KH, Huang Y (2009) Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J Lipid Res 50(Suppl):S183–S188
McWalter GK, Higgins LG, McLellan LI, Henderson CJ, Song L, Thornalley PJ, Itoh K, Yamamoto M, Hayes JD (2004) Transcription factor Nrf2 is essential for induction of NAD(P)H:quinone oxidoreductase 1, glutathione S-transferases, and glutamate cysteine ligase by broccoli seeds and isothiocyanates. J Nutr 134(12 Suppl):3499S–3506S
Miles AT, Hawksworth GM, Beattie JH, Rodilla V (2000) Induction, regulation, degradation, and biological significance of mammalian metallothioneins. Crit Rev Biochem Mol Biol 35(1):35–70
Mocchegiani E, Costarelli L, Giacconi R, Piacenza F, Basso A, Malavolta M (2011) Zinc, metallothioneins and immunosenescence: effect of zinc supply as nutrigenomic approach. Biogerontology. doi:10.1007/s10522-10011-19337-10524
Newman TC, Dawson PA, Rudel LL, Williams DL (1985) Quantitation of apolipoprotein E mRNA in the liver and peripheral tissues of nonhuman primates. J Biol Chem 260(4):2452–2457
Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M (2008) Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem 283(48):33554–33562
Palmiter RD (1987) Molecular biology of metallothionein gene expression. Experientia Suppl 52:63–80
Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W (1993) Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J 12(4):1355–1362
Rimbach G, Pallauf J (1997) Cadmium accumulation, zinc status, and mineral bioavailability of growing rats fed diets high in zinc with increasing amounts of phytic acid. Biol Trace Elem Res 57(1):59–70
Rungapamestry V, Duncan AJ, Fuller Z, Ratcliffe B (2006) Changes in glucosinolate concentrations, myrosinase activity, and production of metabolites of glucosinolates in cabbage (Brassica oleracea Var. capitata) cooked for different durations. J Agric Food Chem 54(20):7628–7634
Schachter F, Faure-Delanef L, Guenot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D (1994) Genetic associations with human longevity at the APOE and ACE loci. Nat Genet 6(1):29–32
Stephens JW, Bain SC, Humphries SE (2008) Gene-environment interaction and oxidative stress in cardiovascular disease. Atherosclerosis 200(2):229–238
Swindell WR (2011) Metallothionein and the biology of aging. Ageing Res Rev 10(1):132–145
Tang L, Zhang Y (2004) Dietary isothiocyanates inhibit the growth of human bladder carcinoma cells. J Nutr 134(8):2004–2010
Venugopal R, Jaiswal AK (1996) Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci USA 93(25):14960–14965
Wagner AE, Ernst I, Iori R, Desel C, Rimbach G (2010) Sulforaphane but not ascorbigen, indole-3-carbinole and ascorbic acid activates the transcription factor Nrf2 and induces phase-2 and antioxidant enzymes in human keratinocytes in culture. Exp Dermatol 19(2):137–144
Wagner AE, Boesch-Saadatmandi C, Dose J, Schultheiss G, Rimbach G (2011) Anti-inflammatory potential of allyl-isothiocyanate-role of Nrf2, NFkappaB and microRNA-155. J Cell Mol Med. doi:10.1111/j.1582-4934.2011.01367.x
Weisgraber KH, Rall Jr SC, Mahley RW (1981) Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem 256(17):9077–9083
Williams DL, Dawson PA, Newman TC, Rudel LL (1985) Apolipoprotein E synthesis in peripheral tissues of nonhuman primates. J Biol Chem 260(4):2444–2451
Yang X, Doser TA, Fang CX, Nunn JM, Janardhanan R, Zhu M, Sreejayan N, Quinn MT, Ren J (2006) Metallothionein prolongs survival and antagonizes senescence-associated cardiomyocyte diastolic dysfunction: role of oxidative stress. Faseb J 20(7):1024–1026
Ye L, Zhang Y (2001) Total intracellular accumulation levels of dietary isothiocyanates determine their activity in elevation of cellular glutathione and induction of Phase 2 detoxification enzymes. Carcinogenesis 22(12):1987–1992
Yeh CT, Yen GC (2005) Effect of sulforaphane on metallothionein expression and induction of apoptosis in human hepatoma HepG2 cells. Carcinogenesis 26(12):2138–2148
Zhang Y, Talalay P (1998) Mechanism of differential potencies of isothiocyanates as inducers of anticarcinogenic Phase 2 enzymes. Cancer Res 58(20):4632–4639
Zhang Y, Li J, Tang L (2005) Cancer-preventive isothiocyanates: dichotomous modulators of oxidative stress. Free Radic Biol Med 38(1):70–77
Zhang YK, Yeager RL, Tanaka Y, Klaassen CD (2010) Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine- and choline-deficient diet. Toxicol Appl Pharmacol 245(3):326–334
Acknowledgments
We thank the German Ministry of Education and Science (BMBF 0313856A, BMBF 0315397C) and the DFG Cluster of Excellence ‘Inflammation at Interfaces’ for financial support. A.C.G. is supported by a grant from H. W. Schaumann foundation. We thank Anja Marx for excellent technical assistance.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Graeser, AC., Huebbe, P., Storm, N. et al. Apolipoprotein E genotype affects tissue metallothionein levels: studies in targeted gene replacement mice. Genes Nutr 7, 247–255 (2012). https://doi.org/10.1007/s12263-012-0282-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12263-012-0282-x