Abstract
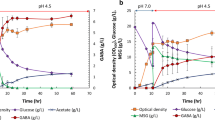
2,3-Butanediol (2,3-BDO) is a promising bio-based chemical for its wide range of applications in industrial areas as synthetic rubber precursor, food additives, and cosmetics. In this study, Escherichia coli DH5α was metabolically engineered for enhanced production of 2,3-BDO by expressing Bacillus subtilis alsS, alsD, and ydjL genes encoding α-acetolactate synthase, α-acetolactate decarboxylase, and acetoin reductase/2,3 -butanediol dehydro¬genase, respectively, along with Deinococcus radiodurans dr1558 gene encoding a response regulator. When recombinant E. coli DH5α strain expressing only B. subtilis alsS, alsD, ydjL genes was cultured in LB medium containing 20 g/L glucose, 3.14 g/L of 2,3-BDO was produced. Additional expression of D. radiodurans dr1558 gene in E. coli DH5α expressing alsS, alsD, and ydjL genes resulted in the production of 7.81 g/L of 2,3-BDO under the same culture conditions, which is 2.5 fold higher than that produced by the strain without DR1558. Transcriptional analysis of E. coli DH5α expressing DR1558 suggested that the expression levels of the genes related to 2,3-BDO pathways were enhanced, while those of genes related to by-product pathways were suppressed, compared with control strain expressing only 2,3-BDO synthesis genes. These results strongly suggest that introduction of the stress tolerant response regulator DR1558 can modulate metabolic pathways to favor production of the target product.
Similar content being viewed by others
References
Jang, Y. S., B. Kim, J. H. Shin, Y. J. Choi, S. Choi, C. W. Song, J. Lee, H. G. Park, and S. Y. Lee (2012) Bio-based production of C2-C6 platform chemicals. Biotechnol. Bioeng. 109: 2437–2459.
Baritugo, K. A., H. T. Kim, Y. David, J. I. Choi, S. H. Hong, K. J. Jeong, J. H. Choi, J. C. Joo, and S. J. Park (2018) Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery. Appl. Microbiol. Biotechnol. 102: 3915–3937.
Oh, Y. H., I. Y. Eom, J. C. Joo, J. H. Yu, B. K. Song, S. H. Lee, S. H. Hong, and S. J. Park (2015) Recent advances in development of biomass pretreatment technologies used in biorefinery for the production of bio-based fuels, chemicals and polymers. Korean J. Chem. Eng. 32: 1945–1959.
Shin, H. Y., S. H. Shim, Y. J. Ryu, J. H. Yang, S. M. Lim, and C. G. Lee (2018) Lipid extraction from Tetraselmis sp. microalgae for biodiesel production using hexane-based solvent mixtures. Biotechnol. Bioprocess Eng. 23: 16–22.
Moon, Y. M., R. Gurav, J. Kim, Y. G. Hong, S. K. Bhatia, H. R. Jung, J. W. Hong, T. R. Choi, S. Y. Yang, H. Y. Park, H. S. Joo, and Y. H. Yang (2018) Whole-cell immobilization of engineered Escherichia coli JY001 with barium-alginate for itaconic acid production. Biotechnol. Bioprocess Eng. 23: 442–447.
Lee, J. W., T. Y. Kim, Y. S. Jang, S. Choi, and S.Y. Lee (2011) Systems metabolic engineering for chemicals and materials. Trends Biotechnol. 29: 370–378.
Becker, J. and C. Wittmann (2015) Advanced biotechnology: Metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew Chem Int Ed Engl. 54: 3328–3350.
Baritugo, K. A. G., H. T. Kim, Y. C. David, J. H. Choi, J. I. Choi, T. W. Kim, C. Park, S. H. Hong, J. G. Na, K. J. Jeong, J. C. Joo, and S. J. Park (2018) Recent advances in metabolic engineering of Corynebacterium glutamicum as a potential platform micro¬organism for biorefinery. Biofuel. Bioprod. Biorefin. 12: 899–925.
Choi, S. Y., S. J. Park, W. J. Kim, J. E. Yang, H. Lee, J. Shin, and S. Y. Lee (2016) One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nat Biotechnol. 34: 435–440.
Joo, J. C., A. N. Khusnutdinova, R. Flick, T. Kim, U. T. Bornscheuer, A. F. Yakunin, and R. Mahadevan (2017) Alkene hydrogenation activity of enoate reductases for an environmentally benign biosynthesis of adipic acid. Chem. Sci. 8: 1406–1413.
Erickson, B., J. E. Nelson, and P. Winters (2012) Perspective on opportunities in industrial biotechnology in renewable chemicals. Biotechnol. J. 7: 176–185.
Ding, S. and T. Tan (2006) L-lactic acid production by Lactobacillus casei fermentation using different fed-batch feeding strategies. Process Biochem. 41: 1451–1454.
Borodina, I., K. R. Kildegaard, N. B. Jensen, T. H. Blicher, J. Maury, S. Sherstyk, K. Schneider, P. Lamosa, M. J. Herrgård, I. Rosenstand, F. Öberg, J. Forster, and J. Nielsen (2015) Establishing a synthetic pathway for high-level production of 3 -hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine. Metab. Eng. 27: 57–64.
Park, S. J., T. W. Kim, M. K. Kim, S. Y. Lee, and S. C. Lim (2012) Advanced bacterial polyhydroxyalkanoates: towards a versatile and sustainable platform for unnatural tailor-made polyesters. Biotechnol. Adv. 30: 1196–1206.
Raab, A. M., G. Gebhardt, N. Bolotina, D. Weuster-Botz, and C. Lang (2010) Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab. Eng. 12: 518–525.
Song, H. and S. Y. Lee (2006) Production of succinic acid by bacterial fermentation. Enzyme Microb. Technol. 39: 352–361.
Garde, A., G. Jonsson, A. S. Schmidt, and B. K. Ahring (2002) Lactic acid production from wheat straw hemicellulose hydrolysate by Lactobacillus pentosus and Lactobacillus brevis. Bioresour Technol. 81: 217–223.
Berezina, O. V., N. V. Zakharova, A. Brandt, S. V. Yarotsky, W. H. Schwarz, and V. V. Zverlov (2010) Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl. Microbiol. Biotechnol. 87: 635–646.
Atsumi, S., T. Y. Wu, E. M. Eckl, S. D. Hawkins, T. Buelter, and J. C. Liao (2010) Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl. Microbiol. Biotechnol. 85: 651–657.
Andreeßen, B., A. B. Lange, H. Robenek, and A. Steinbuchel (2010) Conversion of glycerol to poly (3-hydroxypropionate) in recombinant Escherichia coli. Appl. Environ. Microbiol. 76: 622–626.
Ji, X. J., H. Huang, and P. K. Ouyang (2011) Microbial 2, 3-butanediol production: a state-of-the-art review. Biotechnol. Adv. 29: 351–364.
Winfield, M. E. (1950) The catalytic dehydration of 2, 3-butanediol to butadiene. II. Adsorption equilibria. Aust. J. Chem. 3: 290–305.
Multer, A., N. McGraw, K. Holm, and P. Vadlani (2013) Production of methyl ethyl ketone from biomass using a hybrid biochemical/catalytic approach. Lnd. Eng. Chem. Res. 52: 56–60.
De Faveri, D., P. Torre, F. Molinari, P. Perego, and A. Converti (2003) Carbon material balances and bioenergetics of 2, 3-butanediol bio-oxidation by Acetobacter hansenii. Enzyme Microb. Technol. 33: 708–719.
Xiao, Z. and P. Xu (2007) Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33: 127–140.
Bartowsky, E. J. and P. A. Henschke (2004) The ‘buttery’ attribute of wine—diacetyl—desirability, spoilage and beyond. Int. J. Food Microbiol. 96: 235–252.
Celihska, E. and W. Grajek (2009) Biotechnological production of 2, 3-butanediol—current state and prospects. Biotechnol. Adv. 27: 715–725.
Ng, C. Y., M. Y. Jung, J. Lee, and M. K. Oh (2012) Production of 2, 3-butanediol in Saccharomyces cerevisiae by in silico aided metabolic engineering. Microb. Cell Fact. 11: 68.
Xu, Y., H. Chu, C. Gao, F. Tao, Z. Zhou, K. Li, L. Li, C. Ma, and P. Xu (2014) Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab. Eng. 23: 22–33.
Chu, H., B. Xin, P. Liu, Y. Wang, L. Li, X. Liu, X. Zhang, C. Ma, P. Xu, and C. Gao (2015) Metabolic engineering of Escherichia coli for production of (2S, 3S)-butane-2, 3-diol from glucose. Biotechnol. Biofuels. 8: 143.
Nicholson, W. L. (2008) The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2, 3-butanediol dehydrogenase. Appl. Environ. Microbiol. 74: 6832–6838.
Nicolaou, S. A., S. M. Gaida, and E. T. Papoutsakis (2010) A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 12: 307–331.
Zhang, Y., Y. Zhu, Y. Zhu, and Y. Li (2009) The importance of engineering physiological functionality into microbes. Trends Biotechnol. 27: 664–672.
Chen, T., J. Wang, R. Yang, J. Li, M. Lin, and Z. Lin (2011) Laboratory-evolved mutants of an exogenous global regulator, IrrE from Deinococcus radiodurans, enhance stress tolerances of Escherichia coli. PLoS One. 6: e16228.
Gao, G., B. Tian, L. Liu, D. Sheng, B. Shen, and Y. Hua (2003) Expression of Deinococcus radiodurans PprI enhances the radioresistance of Escherichia coli. DNA Repair. 2: 1419–1427.
Pan, J., J. Wang, Z. Zhou, Y. Yan, W. Zhang, W. Lu, S. Ping, Q. Dai, M. Yuan, B. Feng, X. Hou, Y. Zhang, M. Ruiqiang, T. Liu, L. Feng, L. Wang, M. Chen, and M. Lin (2009) IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS One. 4: e4422.
Ma, R., Y. Zhang, H. Hong, W. Lu, M. Lin, M. Chen, and W. Zhang (2011) Improved osmotic tolerance and ethanol production of ethanologenic Escherichia coli by IrrE, a global regulator of radiation-resistance of Deinococcus radiodurans. Curr Microbiol. 62: 659–664.
Guo, S., X. Yi, W. Zhang, M. Wu, F. Xin, W. Dong, M. Zhang, J. Ma, H. Wu, and M. Jiang (2017) Inducing hyperosmotic stress resistance in succinate-producing Escherichia coli by using the response regulator DR1558 from Deinococcus radiodurans. Process Biochem. 61: 30–37.
Park, S. H., H. Singh, D. Appukuttan, S. Jeong, Y. J. Choi, J. H. Jung, I. Narumi, and S. Lim (2017) PprM, a cold shock domain-containing protein from Deinococcus radiodurans, confers oxidative stress tolerance to Escherichia coli. Front. Microbiol. 7: 2124.
Park, S. H., G. B. Kim, H. U. Kim, S. J. Park, and J. I. Choi (2019) Enhanced production of poly-3-hydroxybutyrate (PHB) by expression of response regulator DR1558 in recombinant Escherichia coli. Int. J. Biol. Macromol. 131: 29–35.
Park, S. J., Y. A. Jang, H. Lee, A. R. Park, J. E. Yang, J. Shin, Y. H. Oh, B. K. Song, J. Jegal, S. H. Lee, and S. Y. Lee (2013) Metabolic engineering of Ralstonia eutropha for the biosynthesis of 2-hydroxyacid-containing polyhydroxyalkanoates. Metab. Eng. 20: 20–28.
Appukuttan, D., H. Singh, S. H. Park, J. H. Jung, S. Jeong, H. S. Seo, Y. J. Choi, and S. Lim (2016) Engineering synthetic multistress tolerance in Escherichia coli by using a Deinococcal response regulator, DR1558. Appl. Environ. Microbiol. 82: 1154–1166.
Sambrook, J. and D. Russell (2001) Molecular Cloning: A Laboratory Manual. 3rd ed.. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY., USA.
Livak, K. J. and T. D. Schmittgen (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2—ΔΔCT method. Methods. 25: 402–408.
Luli, G. W. and W. R. Strohl (1990) Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56: 1004–1011.
Farmer, W. R. and J. C. Liao (1997) Reduction of aerobic acetate production by Escherichia coli. Appl. Environ. Microbiol. 63: 3205–3210.
Siddiquee, K. A., M. J. Arauzo-Bravo, and K. Shimiz (2004) Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fluxes in Escherichia coli. FEMS Microbiol. Lett. 235: 25–33.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2018R1D1A1B07049359), the Golden Seed Project Grant funded by Ministry of Oceans and Fisheries (213008-05-3-SB910), the Ewha Womans University Research Grant of 2017, and the supporting program by Chonnam Nation University (2019-3367).
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Materials
12257_2019_306_MOESM1_ESM.pdf
Enhanced Production of 2,3-Butanediol in Recombinant Escherichia coli Using Response Regulator DR1558 Derived from Deinococcus radiodurans
Rights and permissions
About this article
Cite this article
Park, SJ., Sohn, Y.J., Park, S.J. et al. Enhanced Production of 2,3-Butanediol in Recombinant Escherichia coli Using Response Regulator DR1558 Derived from Deinococcus radiodurans. Biotechnol Bioproc E 25, 45–52 (2020). https://doi.org/10.1007/s12257-019-0306-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-019-0306-0