Abstract
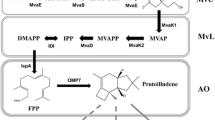
The biosynthesis of terpenoids in heterologous hosts has become increasingly popular. Isopentenyl diphosphate (IPP) is the central precursor of all isoprenoids, and the synthesis can proceed via two separate pathways in different organisms: The 1-deoxylulose 5-phosphate (DXP) pathway and the mevalonate (MVA) pathway. In this study, an in silico comparison was made between the maximum theoretical IPP yields and the thermodynamic properties of the DXP and MVA pathways using different hosts and carbon sources. We found that Escherichia coli and its DXP pathway have the most potential for IPP production. Consequently, codon usage redesign, and combinations of chromosomal engineering and various strains were considered for optimizing taxadiene biosynthesis through the endogenic DXP pathway. A high production strain yielding 876 ± 60 mg/L taxadiene, with an overall volumetric productivity of 8.9 mg/(L × h), was successfully obtained by combining the chromosomal engineered upstream DXP pathway and the downstream taxadiene biosynthesis pathway. This is the highest yield thus far reported for taxadiene production in a heterologous host. These results indicate that genetic manipulation of the DXP pathway has great potential to be used for production of terpenoids, and that chromosomal engineering is a powerful tool for heterologous biosynthesis of natural products.
Similar content being viewed by others
References
Wani, M. C., H. L. Taylor, M. E. Wall, P. Coggon, and A. T. McPhail (1971) Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 93: 2325–2327.
Horwitz, S. B. (1994) How to make taxol from scratch. Nature 367: 593–594.
Frense, D. (2007) Taxanes: Perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 73: 1233–1240.
Roberts, S. C. (2007) Production and engineering of terpenoids in plant cell culture. Nature Chem. Biol. 3: 387–395.
Nicolaou, K. C., Z. Yang, J. J. Liu, H. Ueno, P. G. Nantermet, R. K. Guy, C. F. Claiborne, J. Renaud, E. A. Couladouros, and K. Paulvannan (1994) Total synthesis of taxol. Nature 367: 630–634.
Kirby, J. and J. D. Keasling (2009) Biosynthesis of plant isoprenoids: Perspectives for microbial engineering. Annu. Rev. Plant Biol. 60: 335–355.
Ro, D. K., E. M. Paradise, M. Ouellet, K. J. Fisher, K. L. Newman, J. M. Ndungu, K. A. Ho, R. A. Eachus, T. S. Ham, J. Kirby, M. C. Chang, S. T. Withers, Y. Shiba, R. Sarpong, and J. D. Keasling (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943.
Chang, M. C. and J. D. Keasling (2006) Production of isoprenoid pharmaceuticals by engineered microbes. Nat. Chem. Biol. 2: 674–681.
Martin, V. J., D. J. Pitera, S. T. Withers, J. D. Newman, and J. D. Keasling (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21: 796–802.
Tsuruta, H., C. J. Paddon, D. Eng, J. R. Lenihan, T. Horning, L. C. Anthony, R. Regentin, J. D. Keasling, N. S. Renninger, and J. D. Newman (2009) High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS One. 4: e4489.
Morrone, D., L. Lowry, M. K. Determan, D. M. Hershey, M. Xu, and R. J. Peters (2010) Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 85: 1893–1906.
Huang, Q., C. A. Roessner, R. Croteau, and A. I. Scott (2001) Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg. Med. Chem. 9: 2237–2242.
Besumbes, O., S. Sauret-Gueto, M. A. Phillips, S. Imperial, M. Rodriguez-Concepcion, and A. Boronat (2004) Metabolic engineering of isoprenoid biosynthesis in Arabidopsis for the production of taxadiene, the first committed precursor of Taxol. Biotechnol. Bioeng. 88: 168–175.
Dejong, J. M., Y. Liu, A. P. Bollon, R. M. Long, S. Jennewein, D. Williams, and R. B. Croteau (2006) Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol. Bioeng. 93: 212–224.
Engels, B., P. Dahm, and S. Jennewein (2008) Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 10: 201–206.
Yuan, L. Z., P. E. Rouviere, R. A. Larossa, and W. Suh (2006) Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab. Eng. 8: 79–90.
Jin, Y. S. and G. Stephanopoulos (2007) Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab. Eng. 9: 337–347.
Alper, H., K. Miyaoku, and G. Stephanopoulos (2005) Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23: 612–616.
Feist, A. M., C. S. Henry, J. L. Reed, M. Krummenacker, A. R. Joyce, P. D. Karp, L. J. Broadbelt, V. Hatzimanikatis, and B. O. Palsson (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3: 121.
Herrgard, M. J., N. Swainston, P. Dobson, W. B. Dunn, K. Y. Arga, M. Arvas, N. Bluthgen, S. Borger, R. Costenoble, M. Heinemann, M. Hucka, N. Le Novere, P. Li, W. Liebermeister, M. L. Mo, A. P. Oliveira, D. Petranovic, S. Pettifer, E. Simeonidis, K. Smallbone, I. Spasic, D. Weichart, R. Brent, D. S. Broomhead, H. V. Westerhoff, B. Kirdar, M. Penttila, E. Klipp, B. O. Palsson, U. Sauer, S. G. Oliver, P. Mendes, J. Nielsen, and D. B. Kell (2008) A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 26: 1155–1160.
Oh, Y. K., B. O. Palsson, S. M. Park, C. H. Schilling, and R. Mahadevan (2007) Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J. Biol. Chem. 282: 28791–28799.
Becker, S. A., A. M. Feist, M. L. Mo, G. Hannum, B. O. Palsson, and M. J. Herrgard (2007) Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox. Nat. Protoc. 2: 727–738.
Varma, A. and B. O. Palsson (1993) Metabolic capabilities of Escherichia coli: I. Synthesis of biosynthetic precursors and cofactors. J. Theor. Biol. 165: 477–502.
Varmar, A. and B. O. Palsson (1993) Metabolic capabilities of Escherichia-coli: 2. Optimal-growth patterns. J. Theor. Biol. 165: 503–522.
Gonzalez-Lergier, J., L. J. Broadbelt, and V. Hatzimanikatis (2006) Analysis of the maximum theoretical yield for the synthesis of erythromycin precursors in Escherichia coli. Biotechnol. Bioeng. 95: 638–644.
Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA. 97: 5978–5983.
Datsenko, K. A. and B. L. Wanner (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 97: 6640–6645.
Wang, Y. and B. A. Pfeifer (2008) 6-deoxyerythronolide B production through chromosomal localization of the deoxyerythronolide B synthase genes in E. coli. Metab. Eng. 10: 33–38.
Cunningham, F. X. Jr., Z. Sun, D. Chamovitz, J. Hirschberg, and E. Gantt (1994) Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell. 6: 1107–1121.
Sambrook, J. and D. W. Russell (2001) Molecular cloning: A laboratory manual. 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Lau, J., C. Tran, P. Licari, and J. Galazzo (2004) Development of a high cell-density fed-batch bioprocess for the heterologous production of 6-deoxyerythronolide B in Escherichia coli. J. Biotechnol. 110: 95–103.
Pfeifer, B., Z. Hu, P. Licari, and C. Khosla (2002) Process and metabolic strategies for improved production of Escherichia coliderived 6-deoxyerythronolide B. Appl. Env. Microbiol. 68: 3287–3292.
Jones, K. L., S. W. Kim, and J. D. Keasling (2000) Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab. Eng. 2: 328–338.
Edwards, J. S. and B. O. Palsson (2000) Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics 1: 1.
Segre, D., D. Vitkup, and G. M. Church (2002) Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA. 99: 15112–15117.
Alper, H. and G. Stephanopoulos (2008) Uncovering the gene knockout landscape for improved lycopene production in E. coli. Appl. Microbiol. Biotechnol. 78: 801–810.
Park, J. H., K. H. Lee, T. Y. Kim, and S. Y. Lee (2007) Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proc. Natl. Acad. Sci. USA. 104: 7797–7802.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meng, H., Wang, Y., Hua, Q. et al. In silico analysis and experimental improvement of taxadiene heterologous biosynthesis in Escherichia coli. Biotechnol Bioproc E 16, 205–215 (2011). https://doi.org/10.1007/s12257-010-0329-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-010-0329-z