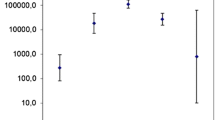
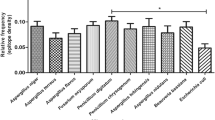
Abstract
Protein therapeutics, particularly of heterologous origin are shown to elicit immunogenic responses which result in adverse allergic reactions in spite of their promising clinical benefit. L-Asparaginase is one such well known chemotherapeutic agent that has enhanced the survival rates to 90 % in the treatment of acute lymphoblastic leukaemia for past 30 years. But the use of this enzyme is accompanied by hypersensitive reactions ranging from allergy to anaphylactic shock which have a drastic influence in treatment outcomes. Numerous attempts have been made to minimize the problems of immunogenicity, which remained as a major bottleneck in the treatment protocols. Conjugating the enzyme L- Asparaginase with PEG was successful as it has reduced the complications in therapy and frequency of injections (dosages), and thus became prominent in reducing the immunogenicity up to a certain extent. Keeping the bottlenecks in consideration during the development of therapeutics, the present study concentrates on engineering of protein as an alternative to the PEGylated enzyme, having reduced immunogenicity as an inbuilt character of protein by using in silico approaches. L-Asparaginase from Escherichia coli and Pectobacterium carotovorum were selected for the present study. The methodology consists of (i) locating the B and CD4+ T cell epitopes of enzyme by in silico tools (ii) generating point mutations of these epitopes to alter or reduce the immunogenicity of protein (iii) generating enzyme models by molecular modelling (iv) assessing the binding affinity of the substrate with L-Asparaginase variants by in silico docking methods using Autodock 4.2 and (v) validating the mutated model for stability by molecular dynamics simulation studies using Gromacs.














Similar content being viewed by others
References
De Duve C (1966) The significance of lysosome in pathology and medicine. Proc Inst Med Chic 26:73–76
Offman MN, Krol M, Patel N, Krishnan S, Liu JZ, Saha V, Bates PA (2011) Rational engineering of L- asparaginase reveals importance of dual activity for cancer cell toxicity. Blood 117:1614–1621
Wai Kin Chan, Philip L. Lorenzi, Andriy Anishkin, Preeti Purwaha, David M. Rogers, Sergei, Sukharev, Susan B. Rempe, and John N. Weinstein. (2014). The glutaminase activity of L-asparaginase is not required for anticancer activity against ASNS-negative cells. Blood. doi: 10.1182/blood-2013-10-535112.
Stams WAG, den Boer ML, Holleman A, Appel IM, Beverloo B, van Wering ER, Janka-Schaub GE, Evans WE, Pieters R (2005) Asparagine synthetase expression is linked with l-asparaginase resistance in TEL-AML1-negative but not TEL-AML1-positive pediatric acute lymphoblastic leukemia. Blood 105:4223–4225
Narta UK, Kanwar SS, Azmi W (2007) Pharmacological and clinical evaluation of L-asparaginase in the treatment if leukaemia. Crit Rev Oncol/Hematol 61:208–221
Pieters R, Hunger S, Boos J et al (2011) L-asparaginase treatment in acute lymphoblastic leukemia. Cancer 117:238–249
Avramis VI, Sencer S, Periclou AP et al (2002) A randomized comparison of native escherichia coli asparaginase and polyethylene glycol conjugated asparaginase for treatment of children with newly diagnosed standard-risk acute lymphoblastic leukemia: a children’s cancer group study. Blood 99:1986–1994
Zalewska-Szewczyk B, Gach A, Wyka K et al (2009) The cross-reactivity of antiasparaginase antibodies against different L-asparaginase preparations. Clin Exp Med 9:113–116
Raetz EA, Salzer WL (2010) Tolerability and efficacy of L-asparaginase therapy in pediatric patients with acute lymphoblastic leukemia. J Pediatr Oncol 32:554–563
Rizzari C, Citterio M, Zucchetti M et al (2006) A pharmacological study on pegylated asparaginase used in front-line treatment of children with acute lymphoblastic leukemia. Haematologica 91:24–31
Abuchowski A, Van Es T, Palczuk NC (1979) Treatment of L5178Y tumor bearing BDf mice with a non-immunogenic l-glutaminaseasparaginase. Cancer Treat Rep 63:1127–1129
Hawkins DS, Park JR, Thomson BG et al (2004) Asparaginase pharmacokinetics after intensive polyethylene glycol-conjugated L-asparaginase therapy for children with relapsed acute lymphoblastic leukemia. Clin Cancer Res 10:5335–5341
Albertsen BK, Schrøder H, Jakobsen P et al (2002) Antibody formation during intravenous and intramuscular therapy with Erwinia asparaginase. Med Pediatr Oncol 38:310–316
Klug Albertsen B, Schmiegelow K, Schrøder H et al (2002) Anti-Erwinia asparaginase antibodies during treatment of childhood acute lymphoblastic leukemia and their relationship to outcome: a case–control study. Cancer Chemother Pharmacol 50:117–120
Vrooman LM, Supko JG, Neuberg DS et al (2010) Erwinia asparaginase after allergy to E. coli asparaginase in children with acute lymphoblastic leukemia. Pediatr Blood Cancer 54:199–205
Kumar GT, Prakash NJ, Laxmi N, Krishna TD (2011) Biotechnology based durg delivery by PEGylation method. IJRAP 2(1):95–102
Kumar S, Pakshirajan K, Venkata Dasu V (2009) Development of medium for enhanced production of glutaminase-free L-asparaginase from Pectobacterium carotovorum MTCC 1428. Appl Microbiol Biotechnol 84:477–486
Kumar S, Pakshirajan K, Venkata Dasu V (2010) Assessment of physical process conditions for enhanced production of novel glutaminase-free L-Asparaginase from Pectobacterium carotovorum MTCC 1428. Appl Biochem Biotechnol 163:327–337
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2001) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Bailey T L, Elkan C (1994) “Fitting a mixture model by expectation maximization to discover motifs in biopolymers,” in Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology. AAAI Press, pp 28–36
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD (2012) Nucleic acids research. Database Issue 40:D290–D301
Zhang Q, Wang P, Kim Y (2008) Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res 36(Web Server issue):W513–W518
Emini EA, Hughes JV, Perlow DS, Boger J (1985) Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol 55:836–839, PMID: 2991600
Parker JM, Guo D, Hodges RS (1986) New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with immunogenicity and X-ray-derived accessible sites. Biochemistry 25:5425–5432, PMID: 2430611
Nielsen M, Lundegaard C, Lund O (2007) Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinf 8:238
Cantor JR, Yoo TH, Dixit A, Iverson BL, Forsthuber TG, Georgiou G (2011) Therapeutic enzyme deimmunization by combinatorial T-cell epitope removal using neutral drift. PNAS 108(4):1272–1277
Hete’nyi C, Spoel VD (2002) Protein Sci 11:1729 [PMID:12070326]
Sousa SF, Fernandes PA, Ramos MJ et al (2006) Proteins 65:15, PMID: 16862531
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semi empirical free energy force field with charge-based desolvation. J Comput Chem 28:145–152
Wang B, Relling MV, Storm MC et al (2003) Evaluation of immunologic crossreaction of antiasparaginase antibodies in acute lymphoblastic leukemia (ALL) and lymphoma patients. Leukemia 17:1583–1588
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Erik L, Berk H, van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 7:306–317
Van der Spoel D, Lindahl E, Hess B, van Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM, Feenstra KA, van Drunen R, Berendsen HJC (2005) Gromacs User Manual version 4.0. www.gromacs.org
Schüttelkopf AW, van Aalten DMF (2004) PRODRG - a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D60:1355–1363
Kaushik Ramakrishanan S, Krishna V, Kumar SV, Lakshmi BS, Anishetty S, Gautham P (2008) Molecular dynamics simulation of lipases. Int J Integr Biol 2:204–213
Clementi A (1922) Desamidation enzymatique de l’ L-Asparagine. Archives Internationales de Physiologie 19:369
Ramya LN, Mukesh D, Rekha VPB, Pulicherla KK (2012) L-Asparaginase as potent anti-leukemic agent and its significance of having reduced glutaminase side activity for the better treatment of acute lymphoblastic leukaemia. Appl Biochem Biotechnol 167(8):2144–2159
Kidd JG (1953) Regression of transplanted lymphomas induced in vivo by means of normal guinea pig serum—course of transplanted cancers of various kinds in mice and rats given guinea pig serum or rabbit serum. J Exp Med 98:565–582
Wade HE, Elsworth R, Herbert D (1968) A new L-asparaginase with anti-tumor activity. Lancet 2:776–777
Manna S, Sinha A, Sadhukhan R, Chakrabarty SL (1995) Purification, characterization and antitumor activity of L-asparaginase isolated from Pseudomonas stutzeri MB-405. Curr Microbiol 30:291–298
Bander NH et al (2005) Phase I trial of 177lutetium-labeled J591, a monoclonal antibody to prostate-specific membrane antigen, in patients with androgen-independent prostate cancer. J Clin Oncol 23:4591–4601
Holgate RG, Baker MP (2009) Circumventing immunogenicity in the development of therapeutic antibodies. IDrugs 12:233–237
Macfarlane DJ et al (2006) Safety, pharmacokinetic and dosimetry evaluation of the proposed thrombus imaging agent 99mTc-DI-DD-3B6/22-80B3 Fab 0. Eur J Nucl Med Mol Imaging 33:648–656
Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4(4):e1000048
Wang P et al (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4:e1000048
Moola ZB, Scawen MD, Atkinson T, Nicholls DJ (1994) Erwinia chrysanthemi L-asparaginase: epitope mapping and production of antigenically modified enzymes. Biochem J 302:921–927
Jianhua C, Yujun W, Ruibo J, Min W, Wutong W (2006) Probing the antigenicity of E. coli L-asparaginase by mutational analysis. Mol Biotechnol 33(1):57–65
Ramya LN, Doble M, Rekha VPB, Pulicherla KK (2011) In silico engineering of L-Asparaginase to have reduced glutaminase side activity for effective treatment of acute lymphoblastic leukaemia. J Pediatr Hematol Oncol 33(8):617–621
Derst C, Henseling J, Rohm KH (1992) Probing the role of threonine and serine residues of E.coli asparaginase II by site-specific mutagenesis. Protein Eng 5:785–789, PubMed: 1287659
Patil R, Das S, Stanley A et al (2010) Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS ONE 5(8):1–10
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ramya, L.N., Pulicherla, K.K. Studies on Deimmunization of Antileukaemic L-Asparaginase to have Reduced Clinical Immunogenicity- An in silico Approach. Pathol. Oncol. Res. 21, 909–920 (2015). https://doi.org/10.1007/s12253-015-9912-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-015-9912-0