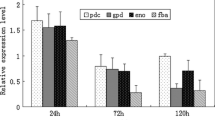
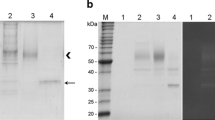
Abstract
Xylanase III (Xyn III), a specific endoxylanase that belongs to family 10 of the glycoside hydrolases, was overexpressed in Trichoderma reesei QM9414 using a constitutive strong promoter of the gene encoding pyruvate decarboxylase (pdc). The maximum recombinant xylanase activity achieved was 817.2 ± 65.2 U/mL in the transformant fermentation liquid. The productivities of Xyn III accounted for approximately 53 % of the total protein secreted by the recombinant. The enzyme was optimally active at 60 °C and pH 6. The recombinant Xyn III was stable at pH 5–8. This is the first report on the homologous expression of xyn3 in T. reesei QM9414. The properties of Xyn III make it promising in a variety of industrial use.



Similar content being viewed by others
References
Akel E, Metz B, Seiboth B, Kubicek CP (2009) Molecular regulation of arabinan and l-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei). Eukaryot Cell 8(12):1837–1844
Bailey MJ, Biely P, Poutanen KJ (1992) Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 23:257–270
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cai HY, Shi PJ, Bai YG, Huang HQ, Yuan TZ, Yang PL, Luo HY, Meng K, Yao B (2011) A novel thermoacidophilic family 10 xylanase from Penicillium pinophilum C1. Process Biochem 46(12):2341–2346
Do TT, Quyen DT, Nguyen TN, Nguyen VT. Molecular characterization of a glycosyl hydrolase family 10 xylanase from Aspergillus niger. Protein Expr Purif. 2013
Du Y, Shi P, Huang H, Zhang X, Luo H, Wang Y, Yao B (2013) Characterization of three novel thermophilic xylanases from Humicola insolens Y1 with application potentials in the brewing industry. Bioresour Technol 130:161–167
Fedorova TV, Chulkin AM, Vavilova EA, Maisuradze IG, Trofimov AA, Zorov IN, Khotchenkov VP, Polyakov KM, Benevolensky SV, Koroleva OV, Lamzin VS (2012) Purification, biochemical characterization, and structure of recombinant endo-1, 4-β-xylanase XylE. Biochemistry (Mosc) 77(10):1190–1198
Fu XY, Zhao W, Xiong AS, Tian YS, Peng RH (2011) High expression of recombinant Streptomyces sp. S38 xylanase in Pichia pastoris by codon optimization and analysis of its biochemical properties. Mol Biol Rep 38(8):4991–4997
Furukawa T, Shida Y, Kitagami N, Ota Y, Adachi M, Nakagawa S, Shimada R, Kato M, Kobayashi T, Okada H, Ogasawara W, Morikawa Y (2008) Identification of the cis-acting elements involved in regulation of xylanase III gene expression in Trichoderma reesei PC-3-7. Fungal Genet Biol 45(7):1002–1094
Gilkes NR, Henrissat B, Kilburn DG, Miller RC Jr, Warren RAJ (1991) Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev 55:303–315
Henrissat B, Claeyssens M, Tomme P, Lemesle L, Mornon JP (1989) Cellulase families revealed by hydrophobic analysis. Gene 81(1):83–95
Huy ND, Kim SW, Park SM (2011) Heterologous expression of endo-1, 4-beta-xylanaseC from Phanerochaete chrysosporium in Pichia pastoris. J Biosci Bioeng 111(6):654–657
Kawamori M, Morikawa Y, Takasawa S (1986) Induction and production of cellulases by l-sorbose in Trichoderma reesei. Appl Microbiol Biotechnol 24:449–453
Li J, Wang J, Wang S, Xing M, Yu S, Liu G (2012) Achieving efficient protein expression in Trichoderma reesei by using strong constitutive promoters. Microb Cell Fact 11:84–93
Mach-Aigner AR, Pucher ME, Steiger MG, Bauer GE, Preis SJ, Mach RL (2008) Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl Environ Microbiol 74(21):6554–6562
Mandels M, Andreotti RE (1978) Problems and changes in the cellulose to cellulase fermentation. Process Biochem 13:6–13
Miller L (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Ogasawara W, Shida Y, Furukawa T, Shimada R, Nakagawa S, Kawamura M, Yagyu T, Kosuge A, Xu J, Nogawa M, Okada H, Morikawa Y (2006) Cloning, functional expression and promoter analysis of xylanase III gene from Trichoderma reesei. Appl Microbiol Biotechnol 72(5):995–1003
Penttilä M, Nevalainen H, Rättö M, Salminen E, Knowles J (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164
Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, Van den HCA (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117–124
Wakiyama M, Tanaka H, Yoshihara K, Hayashi S, Ohta K (2008) Purification and properties of family-10 endo-1, 4-beta-xylanase from Penicillium citrinum and structural organization of encoding gene. J Biosci Bioeng 105(4):367–374
Wang J, Mai GQ, Liu G, Yu SW (2013a) Molecular cloning and heterologous expression of an acid-stable endoxylanase gene from Penicillium oxalicum in Trichoderma reesei. J Microbiol Biotechnol 23(2):251–259
Wang JQ, Yin X, Wu MC, Zhang HM, Gao SJ, Wei JT, Tang CD, Li JF (2013b) Expression of a family 10 xylanase gene from Aspergillus usamii E001 in Pichia pastoris and characterization of the recombinant enzyme. J Ind Microbiol Biotechnol 40(1):75–83
Shin K, Jeya M, Lee JK, Kim YS (2010) Purification and characterization of a thermostable xylanase from Fomitopsis pinicola. J Microbiol Biotechnol 20(10):1415–1423
Xu J, Takakuwa N, Nogawa M, Okada H, Morikawa Y (1998) A third xylanase from Trichoderma reesei PC-3-7. Appl Microbiol Biotechnol 49:718–724
Xu J, Nogawa M, Okada H, Morikawa Y (2000) Regulation of xyn3 gene expression in Trichoderma reesei PC-3-7. Appl Microbiol Biotechnol 54(3):370–375
Zheng J, Guo N, Wu L, Tian J, Zhou H (2013) Characterization and constitutive expression of a novel endo-1, 4-β-d-xylanohydrolase from Aspergillus niger in Pichia pastoris. Biotechnol Lett 5(9):1433–1440
Acknowledgments
This work is supported by the Shenzhen Municipal Science and Technology Basic Research Program (JC201005280398A) and Shenzhen Municipal Science and Technology key projects of the Basic Research Program (JC201005250041A).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, J., Zeng, D., Mai, G. et al. Homologous constitutive expression of Xyn III in Trichoderma reesei QM9414 and its characterization. Folia Microbiol 59, 229–233 (2014). https://doi.org/10.1007/s12223-013-0288-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-013-0288-9