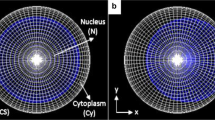
Abstract
Objective
Cellular dosimetry plays a crucial role in radiobiology and evaluation of the relative merits of radiopharmaceuticals used for targeted radionuclide therapy. The present study aims to investigate the effects of various cell geometries on dosimetric characteristics of several Auger emitters distributed in different subcellular compartments using Monte Carlo simulation.
Methods
The Geant4-DNA extension of the Geant4 Monte Carlo simulation toolkit was employed to calculate the mean absorbed dose per unit cumulated activity (S value) for different subcellular distributions of several Auger electron-emitting theranostic radionuclides including 99mTc, 111In, 123I, 125I, and 201Tl. The simulations were carried out in various single-cell models of liquid water including spherical, ellipsoidal, spherical spindle, and ellipsoidal spindle cell models. The latter two models which are generalized from the first two models were inspired by the morphologies of spindle-shaped (fusiform) cells, and were developed to provide more realistic modeling of this common geometry observed in many healthy and cancerous cells.
Results
Evaluation of the S values calculated for the examined cell models reveals that the differences are small (less than 9%) for the cell ← cell, cell ← cell surface, and nucleus ← nucleus source–target combinations. However, moderate discrepancies are seen (up to 28%) when the nucleus is considered as the target, as well as the radioactivity is either internalized into the cytoplasm or bound to the cell membrane.
Conclusions
The findings of the present work suggest that the assumption of spherical cell geometry may provide reasonably accurate estimates of the cellular/nuclear dose for the considered Auger emitters, even for spindle-shaped cells. Of course, this approximation should be used with caution for the nucleus ← cytoplasm and nucleus ← cell surface configurations, since the S-value sensitivity to the cell geometry is somewhat significant in these cases.






Similar content being viewed by others
References
Emfietzoglou D, Kostarelos K, Hadjidoukas P, Bousis C, Fotopoulos A, Pathak A, et al. Subcellular S-factors for low-energy electrons: a comparison of Monte Carlo simulations and continuous-slowing-down calculations. Int J Radiat Biol. 2008;84:1034–44. https://doi.org/10.1080/09553000802460180.
Santos-Cuevas CL, Ferro-Flores G, Rojas-Calderón EL, García-Becerra R, Ordaz-Rosado D, Arteaga de Murphy C, et al. 99mTc-N2S2-Tat (49–57)-bombesin internalized in nuclei of prostate and breast cancer cells: kinetics, dosimetry and effect on cellular proliferation. Nucl Med Commun. 2011;32:303–13. https://doi.org/10.1097/MNM.0b013e328341b27f.
André T, Morini F, Karamitros M, Delorme R, Le Loirec C, Campos L, et al. Comparison of Geant4-DNA simulation of S-values with other Monte Carlo codes. Nucl Instrum Methods Phys Res B. 2014;319:87–94. https://doi.org/10.1016/j.nimb.2013.11.005.
Emfietzoglou D, Bousis C, Hindorf C, Fotopoulos A, Pathak A, Kostarelos K. A Monte Carlo study of energy deposition at the sub-cellular level for application to targeted radionuclide therapy with low-energy electron emitters. Nucl Instrum Methods Phys Res B. 2007;256:547–53. https://doi.org/10.1016/j.nimb.2006.12.055.
Bousis C, Emfietzoglou D, Hadjidoukas P, Nikjoo H. A Monte Carlo study of cellular S-factors for 1 keV to 1 MeV electrons. Phys Med Biol. 2009;54:5023–38. https://doi.org/10.1088/0031-9155/54/16/012.
Cornelissen B, Vallis KA. Targeting the nucleus: an overview of Auger-electron radionuclide therapy. Curr Drug Discov Technol. 2010;7:263–79. https://doi.org/10.2174/157016310793360657.
Rojas-Calderón EL, Torres-García E, Ávila O. Dose per unit cumulated activity (S-values) for e− and beta emitting radionuclides in cancer cell models calculated by Monte Carlo simulation. Appl Radiat Isot. 2014;90:229–33. https://doi.org/10.1016/j.apradiso.2014.04.012.
Gudkov SV, Shilyagina NY, Vodeneev VA, Zvyagin AV. Targeted radionuclide therapy of human tumors. Int J Mol Sci. 2016;17:33. https://doi.org/10.3390/ijms17010033.
Bardiès M, Pihet P. Dosimetry and microdosimetry of targeted radiotherapy. Curr Pharm Des. 2000;6:1469–502. https://doi.org/10.2174/1381612003399176.
Sgouros G. Dosimetry of internal emitters. J Nucl Med. 2005;46:18S–27S.
Champion C, Zanotti-Fregonara P, Hindié E. CELLDOSE: a Monte Carlo code to assess electron dose distribution—S values for 131I in spheres of various sizes. J Nucl Med. 2008;49:151–7. https://doi.org/10.2967/jnumed.107.045179.
Siragusa M, Baiocco G, Fredericia PM, Friedland W, Groesser T, Ottolenghi A, et al. The COOLER code: a novel analytical approach to calculate subcellular energy deposition by internal electron emitters. Radiat Res. 2017;182:204–20. https://doi.org/10.1667/RR14683.1.
Fourie H, Newman R, Slabbert J. Microdosimetry of the Auger electron emitting 123I radionuclide using Geant4-DNA simulations. Phys Med Biol. 2015;60:3333–46. https://doi.org/10.1088/0031-9155/60/8/3333.
Šefl M, Incerti S, Papamichael G, Emfietzoglou D. Calculation of cellular S-values using Geant4-DNA: the effect of cell geometry. Appl Radiat Isot. 2015;104:113–23. https://doi.org/10.1016/j.apradiso.2015.06.027.
Nikjoo H, Emfietzoglou D, Liamsuwan T, Taleei R, Liljequist R, Uehara S. Radiation track, DNA damage and response—a review. Rep Prog Phys. 2016;79:116601. https://doi.org/10.1088/0034-4885/79/11/116601.
Humm JL, Howell RW, Rao DV. Dosimetry of Auger-electron-emitting radionuclides: report no. 3 of AAPM nuclear medicine task group no. 6. Med Phys. 1994;21:1901–15. https://doi.org/10.1118/1.597227.
Ftáčniková S, Böhm R. Monte Carlo calculations of energy deposition on cellular, multicellular and organ level for Auger emitters. Radiat Prot Dosim. 2000;92:279–88. https://doi.org/10.1093/oxfordjournals.rpd.a033293.
Goddu SM, Howell RW, Bouchet L, Bolch W, Rao DV. MIRD cellular S values. Reston: Society of Nuclear Medicine; 1997.
Howell RW, Wessels BW, Leovinger R, In Collaboration with the MIRD Committee, Society of Nuclear Medicine. The MIRD perspective 1999. J Nucl Med. 1999;40:3S–10S.
Incerti S, Kyriakou I, Bernal MA, Bordage MC, Francis Z, Guatelli S, et al. Geant4-DNA example applications for track structure simulations in liquid water: a report from the Geant4-DNA project. Med Phys. 2018;45:e722. https://doi.org/10.1002/mp.13048.
Howell RW, Rao DV, Sastry KSR. Macroscopic dosimetry for radioimmunotherapy: nonuniform activity distributions in solid tumors. Med Phys. 1989;16:66–74. https://doi.org/10.1118/1.596404.
Bousis C, Emfietzoglou D, Nikjoo H. Monte Carlo single-cell dosimetry of I-131, I-125 and I-123 for targeted radioimmunotherapy of B-cell lymphoma. Int J Radiat Biol. 2012;88:908–15. https://doi.org/10.3109/09553002.2012.666004.
Arnaud FX, Paillas S, Pouget JP, Incerti S, Bardiès M, Bordage MC. Complex cell geometry and sources distribution model for Monte Carlo single cell dosimetry with iodine 125 radioimmunotherapy. Nucl Instrum Methods Phys Res B. 2016;366:227–33. https://doi.org/10.1016/j.nimb.2015.11.008.
Nikjoo H, Uehara S, Emfietzoglou D, Cucinotta FA. Track-structure codes in radiation research. Radiat Meas. 2006;41:1052–74. https://doi.org/10.1016/j.radmeas.2006.02.001.
Bousis C, Emfietzoglou D, Hadjidoukas P, Nikjoo H. Monte Carlo single-cell dosimetry of Auger-electron emitting radionuclides. Phys Med Biol. 2010;55:2555–72. https://doi.org/10.1088/0031-9155/55/9/009.
Falzone N, Fernández-Varea JM, Flux G, Vallis KA. Monte Carlo evaluation of Auger electron-emitting theranostic radionuclides. J Nucl Med. 2015;56:1441–6. https://doi.org/10.2967/jnumed.114.153502.
Taborda A, Benabdallah N, Desbrée A. Dosimetry at the sub-cellular scale of Auger-electron emitter Tc-99 m in a mouse single thyroid follicle. Appl Radiat Isot. 2016;108:58–63. https://doi.org/10.1016/j.apradiso.2015.12.010.
Nettleton JS, Lawson RS. Cellular dosimetry of diagnostic radionuclides for spherical and ellipsoidal geometry. Phys Med Biol. 1996;41:1845–54. https://doi.org/10.1088/0031-9155/41/9/018.
Amato E, Lizio D, Baldari S. Absorbed fractions in ellipsoidal volumes for β− radionuclides employed in internal radiotherapy. Phys Med Biol. 2009;54:4171–80. https://doi.org/10.1088/0031-9155/54/13/013.
Amato E, Lizio D, Baldari S. Absorbed fractions for electrons in ellipsoidal volumes. Phys Med Biol. 2011;56:357–65. https://doi.org/10.1088/0031-9155/56/2/005.
Salim R, Taherparvar P. Monte Carlo single-cell dosimetry using Geant4-DNA: the effects of cell nucleus displacement and rotation on cellular S values. Radiat Environ Biophys. 2019;58:353–71. https://doi.org/10.1007/s00411-019-00788-z.
Ikenaga N, Ohuchida K, Mizumoto K, Akagawa S, Fujiwara K, Eguchi D, et al. Pancreatic cancer cells enhance the ability of collagen internalization during epithelial–mesenchymal transition. PLoS ONE. 2012;7:e40434. https://doi.org/10.1371/journal.pone.0040434.
Duclos G, Erlenkämper C, Joanny JF, Silberzan P. Topological defects in confined populations of spindle-shaped cells. Nat Phys. 2017;13:58–62. https://doi.org/10.1038/nphys3876.
Franchi M, Masola V, Bellin G, Onisto M, Karamanos KA, Piperigkou Z. Collagen fiber array of peritumoral stroma influences epithelial-to-mesenchymal transition and invasive potential of mammary cancer cells. J Clin Med. 2019;8:213. https://doi.org/10.3390/jcm8020213.
Human Protein Atlas. The Human Protein Atlas. 2019. https://www.proteinatlas.org/ENSG00000068903-SIRT2/cell#img and https://www.proteinatlas.org/ENSG00000161800-RACGAP1/cell#img. Accessed 14 Apr 2019.
Sechopoulos I, Rogers DWO, Bazalova-Carter M, Bolch WE, Heath EC, McNitt-Gray MF, et al. RECORDS: improved Reporting of montE CarlO RaDiation transport Studies: report of the AAPM Research Committee Task Group 268. Med Phys. 2018;45:e1–5. https://doi.org/10.1002/mp.12702.
Agostinelli S, Allison J, Amako K, Apostolakis J, Araujo H, Arce P, et al. Geant4—a simulation toolkit. Nucl Instrum Methods Phys Res A. 2003;506:250–303. https://doi.org/10.1016/S0168-9002(03)01368-8.
Allison J, Amako K, Apostolakis J, Araujo H, Arce Dubois P, Asai M, et al. Geant4 developments and applications. IEEE Trans Nucl Sci. 2006;53:270–8. https://doi.org/10.1109/TNS.2006.869826.
Allison J, Amako K, Apostolakis J, Arce P, Asai M, Aso T, et al. Recent developments in Geant4. Nucl Instrum Methods Phys Res A. 2016;835:186–225. https://doi.org/10.1016/j.nima.2016.06.125.
Incerti S, Baldacchino G, Bernal MA, Capra R, Champion C, Francis Z, et al. The Geant4-DNA project. Int J Model Simul Sci Comput. 2010;1:157–78. https://doi.org/10.1142/S1793962310000122.
Bernal MA, Bordage MC, Brown JMC, Davídková M, Delage E, El Bitar Z, et al. Track structure modeling in liquid water: a review of the Geant4-DNA very low energy extension of the Geant4 Monte Carlo simulation toolkit. Phys Med. 2015;31:861–74. https://doi.org/10.1016/j.ejmp.2015.10.087.
Geant4 collaboration. Geant4 user’s guide for application developers, release 10.5. 2019. https://geant4.web.cern.ch/support/user_documentation. Accessed 1 Aug 2019.
Incerti S, Ivanchenko A, Karamitros M, Mantero A, Moretto P, Tran HN, et al. Comparison of GEANT4 very low energy cross section models with experimental data in water. Med Phys. 2010;37:4692–708. https://doi.org/10.1118/1.3476457.
Howell RW. Radiation spectra for Auger-electron emitting radionuclides: report no. 2 of AAPM nuclear medicine task group no. 6. Med Phys. 1992;19:1371–83. https://doi.org/10.1118/1.596927.
Vaziri B, Wu H, Dhawan AP, Du P, Howell RW. MIRD pamphlet no. 25: MIRDcell v2.0 software tool for dosimetric analysis of biologic response of multicellular populations. J Nucl Med. 2014;55:1557–64. https://doi.org/10.2967/jnumed.113.131037.
Incerti S, Douglass M, Penfold S, Guatelli S, Bezak E. Review of Geant4-DNA applications for micro and nanoscale simulations. Phys Med. 2016;32:1187–200. https://doi.org/10.1016/j.ejmp.2016.09.007.
Incerti S, Kyriakou I, Bordage MC, Guatelli S, Ivanchenko V, Emfietzoglou D. Track structure simulations of proximity functions in liquid water using the Geant4-DNA toolkit. J Appl Phys. 2019;125:104301. https://doi.org/10.1063/1.5083208.
Kyriakou I, Emfietzoglou D, Ivanchenko V, Bordage MC, Guatelli S, Lazarakis P, et al. Microdosimetry of electrons in liquid water using the low energy models of Geant4. J Appl Phys. 2017;122:024303. https://doi.org/10.1063/1.4992076.
Lampe N, Karamitros M, Breton V, Brown JMC, Kyriakou I, Sakata D, et al. Mechanistic DNA damage simulations in Geant4-DNA part 1: a parameter study in a simplified geometry. Phys Med. 2018;48:135–45. https://doi.org/10.1016/j.ejmp.2018.02.011.
Lampe N, Karamitros M, Breton V, Brown JMC, Sakata D, Sarramia D, et al. Mechanistic DNA damage simulations in Geant4-DNA part 2: electron and proton damage in a bacterial cell. Phys Med. 2018;48:146–55. https://doi.org/10.1016/j.ejmp.2017.12.008.
Michel RB, Castillo ME, Andrews PM, Mattes MJ. In vitro toxicity of A-431 carcinoma cells with antibodies to epidermal growth factor receptor and epithelial glycoprotein-1 conjugated to radionuclides emitting low-energy electrons. Clin Cancer Res. 2004;10:5957–66. https://doi.org/10.1158/1078-0432.CCR-03-0465.
Ocampo-García BE, Santos-Cuevas CL, León-Rodríguez LM, García-Becerra R, Ordaz-Rosado D, Luna-Gutiérrez MA, et al. Design and biological evaluation of 99mTc-N2S2-Tat(49–57)-c(RGDyK): a hybrid radiopharmaceutical for tumors expressing α(v)β(3) integrins. Nucl Med Biol. 2013;40:481–7. https://doi.org/10.1016/j.nucmedbio.2013.01.003.
Pouget JP, Santoro L, Raymond L, Chouin N, Bardiès M, Bascoul-Mollevi C, et al. Cell membrane is a more sensitive target than cytoplasm to dense ionization produced by auger electrons. Radiat Res. 2008;170:192–200. https://doi.org/10.1667/RR1359.1.
Bousis C. Dosimetry on sub-cellular level for intracellular incorporated Auger-electron-emitting radionuclides: a comparison of Monte Carlo simulation and analytic calculations. Radiat Prot Dosim. 2011;143:33–41. https://doi.org/10.1093/rpd/ncq293.
Eckerman KF, Westfall RJ, Ryman JC, Cristy M. Nuclear decay data files of the Dosimetry Research Group. Tennessee: Oak Ridge National Laboratory (ORNL); 1993. https://doi.org/10.2172/10116928.
Funding
The authors received no specific funding for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Salim, R., Taherparvar, P. Cellular S values in spindle-shaped cells: a dosimetry study on more realistic cell geometries using Geant4-DNA Monte Carlo simulation toolkit. Ann Nucl Med 34, 742–756 (2020). https://doi.org/10.1007/s12149-020-01498-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12149-020-01498-z