Abstract
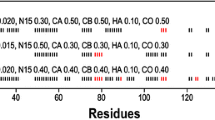
YibK is a tRNA methyltransferase from Haemophilus influenzae, which forms a stable homodimer in solution and contains a deep trefoil 31 knot encompassing the C-terminal helix that threads through a long loop. It has been a model system for investigating knotted protein folding pathways. Recent data have shown that the polypeptide chain of YibK remains loosely knotted under highly denaturing conditions. Here, we report 1H, 13C and 15N chemical shift assignments for YibK and its variant in the presence of 8 M urea. This work forms the basis for further analysis using NMR techniques such as paramagnetic relaxation enhancement, residual dipolar couplings and spin-relaxation dynamics analysis.



Similar content being viewed by others
References
Armengod ME, Moukadiri I, Prado S, Ruiz-Partida R, Benitez-Paez A, Villarroya M, Lomas R, Garzon MJ, Martinez-Zamora A, Meseguer S, Navarro-Gonzalez C (2012) Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie 94:1510–1520
Benitez-Paez A, Villarroya M, Douthwaite S, Gabaldon T, Armengod ME (2010) YibK is the 2′-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors. RNA 16:2131–2143
Cavanagh J (2007) Protein NMR spectroscopy: principles and practice, 2nd edn. Academic Press, Amsterdam
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Goddard TD, Kneller DG SPARKY 3. University of California, San Francisco
Hsieh SJM, Mallam AL, Jackson SE, Hsu STD (2013) 1H, 13C and 15N assignments of YbeA in the 8 M urea-denatured states, Venus. Biomol NMR Assign. doi:10.1007/s12104-013-9501-7
Hsu ST, Behrens C, Cabrita LD, Dobson CM (2009a) 1H, 15N and 13C assignments of yellow fluorescent protein (YFP) Venus. Biomol NMR Assign 3:67–72
Hsu ST, Cabrita LD, Christodoulou J, Dobson CM (2009b) 1H, 15N and 13C assignments of domain 5 of Dictyostelium discoideum gelation factor (ABP-120) in its native and 8 M urea-denatured states. Biomol NMR Assign 3:29–31
Lim K, Zhang H, Tempczyk A, Krajewski W, Bonander N, Toedt J, Howard A, Eisenstein E, Herzberg O (2003) Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a cofactor bound at a site formed by a knot. Proteins 51:56–67
Mallam AL, Jackson SE (2005) Folding studies on a knotted protein. J Mol Biol 346:1409–1421
Mallam AL, Jackson SE (2006) Probing nature’s knots: the folding pathway of a knotted homodimeric protein. J Mol Biol 359:1420–1436
Mallam AL, Jackson SE (2007) The dimerization of an alpha/beta-knotted protein is essential for structure and function. Structure 15:111–122
Mallam AL, Jackson SE (2008) Use of protein engineering techniques to elucidate protein folding pathways. Prog Mol Biol Transl Sci 84:57–113
Mallam AL, Onuoha SC, Grossmann JG, Jackson SE (2008) Knotted fusion proteins reveal unexpected possibilities in protein folding. Mol Cell 30:642–648
Mallam AL, Rogers JM, Jackson SE (2010) Experimental detection of knotted conformations in denatured proteins. Proc Natl Acad Sci USA 107:8189–8194
Acknowledgments
The NMR spectra were obtained at the Core Facility for Protein Structural Analysis supported by National Core Facility Program for Biotechnology. STDH is a recipient of the Career Development Award (CDA—00025/2010-C) from the International Human Frontier Science Program and is supported by funding from the National Science Council (100-2113-M-001-031-MY2 and 101-2627-M-001-004) and Academia Sinica, Taiwan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hsieh, SJ.M., Mallam, A.L., Jackson, S.E. et al. Backbone NMR assignments of a topologically knotted protein in urea-denatured state. Biomol NMR Assign 8, 439–442 (2014). https://doi.org/10.1007/s12104-013-9510-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-013-9510-6