Abstract
Objective
To determine the frequency of respiratory pathogens in infants diagnosed with acute lower respiratory tract infections.
Methods
A prospective cross-sectional observational study was conducted in infants hospitalized with a diagnosis of acute lower respiratory tract infection (ALRTI), in a tertiary care hospital in a metropolitan city of Western India. Nasopharyngeal swabs were analyzed by multiplex real time polymerase chain reaction, for 18 viruses and 3 bacteria (H. influenzae type b, C. pneumoniae and M. pneumoniae). The entire data was entered in Microsoft excel sheet and frequencies were determined.
Results
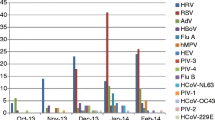
One hundred eligible infants were enrolled. Pathogens were detected in 82 samples, which included Respiratory syncytial viruses (RSV) A / B (35.4%), Human rhinovirus (25.6%), Adenovirus (22%), Human Parainfluenza viruses (11%), Human bocavirus (9.8), Human metapneumovirus A / B (8.5%), Influenza A (H1N1) pdm 09 (6.1%), Parechovirus (3.7%), Human coronaviruses (3.66%), Haemophilus influenzae type b (6.1%), Chlamydia pneumoniae (2.4%) and Mycoplasma pneumoniae (2.4%). Influenza A (other than H1N1), Influenza B, Human Coronavirus 229E and Enterovirus were not detected. The rate of coinfection was 34% and rhinovirus was the most common of the multiple pathogens.
Conclusions
Spectrum of viral etiologies of ALRTI is highlighted. Etiological diagnosis of ALRTI would enable specific antiviral therapy, restrict antibiotic use and help in knowing burden of disease.
Similar content being viewed by others
Introduction
Acute lower respiratory tract infections (ALRTIs) are a leading cause of morbidity and mortality worldwide, especially during the first years of life [1]. They are responsible for 6.8% of deaths in neonates, 20% of deaths in children aged 1–12 mo and 12% of deaths in children aged 1 to 4 y [2] and are considered to be responsible for 22% of deaths in children of age group 0–5 y in Southeast Asia [3]. Respiratory infections are caused by a variety of viruses and bacteria with respiratory viruses being the commonest etiological agent of ALRTI among infants. There is a paucity of data regarding etiological agents for ALRTI among infants from western part of India [4]. Data provided by studies carried out in different parts of India may have some lacunae, because of use of tests which are less sensitive or non-specific or are unable to detect wide range of pathogens simultaneously [5,6,7,8,9]. This data is of importance to ensure that unnecessary prescriptions of anti-bacterial agents are minimized, and appropriate measures are taken to limit the possibility of spread of nosocomial infections. The advent of diagnostic molecular methods (such as multiplex real-time polymerase chain reaction) provides a rare opportunity to identify viruses responsible for respiratory infections as these methods are more sensitive, more accurate and more efficient than traditional methods based on isolation, antibody-detection or serology and are capable of identifying multiple pathogens [1].
Material and Methods
A prospective cross-sectional observational study was carried out in a tertiary care university-affiliated hospital in a metropolitan city in Western India after obtaining approval from the institutional ethics committee. Infants aged 1–12 mo of either gender with ALRTI were enrolled in the study after obtaining written informed consent from the parent or guardian. For the purpose of the study, ALRTI was defined as presence of cough with fever (temperature > 38 °C) for < 2 wk. with tachypnea (50 breaths or more breaths/ min), wheeze, stridor when calm, cyanosis, sub-costal retractions, or apnea [5, 10]. A detailed history was obtained from parent or guardian regarding various risk factors, which were defined in the manner given in Table 1 for the purpose of the study, differential diagnosis provided by the clinician were noted and the information was entered in the Case Record Form. The clinical diagnosis recorded was based on the clinician’s perspective and judgement, which in turn, was made on the basis of information and data obtained from history, clinical examination findings and result of the laboratory and radiological investigation.
Nasopharyngeal secretions were collected using flocked swabs (Puritan Unitranz – RT transport system containing swab and universal transport medium, Puritan Medical Products, USA) by following the standard procedure [14]. Samples were immediately transported to the Molecular diagnostic laboratory, maintaining cold chain & were stored at −80 °C until further use.
Total nucleic acid was manually extracted using an RTP® Pathogen kit (Stratec molecular GmbH, Berlin, Germany) for simultaneous isolation of bacterial & viral DNA, plus viral RNA. Nucleic acid was extracted from 400 μL of universal transport medium containing nasopharyngeal swab (NPS) and was eluted to a final volume of 75 μL according to the RTP ® Pathogen protocol. An internal RNA virus control, the Brome Mosaic Virus (Fast-track Diagnostics, Luxembourg), was introduced into the lysis buffer with each sample, to monitor the nucleic acid extraction and reverse transcription.
Multiplex real time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR):RT- PCR was performed using the FTD Respiratory Pathogen 21 plus (Fast-track Diagnostics, Luxembourg) for detection of Influenza A & B (Inf A & B); Inf A (H1N1) pdm 09; Respiratory syncytial virus (RSV A/B); Human rhinovirus (HRV); Human adenovirus (ADV); Human parainfluenza virus (PIV) 1,2,3,4; Human coronavirus (hCoV) OC43, HKU1, NL63, 229E; Human metapneumovirus (hMPV A/B); Human bocavirus (hBoV A/B); Enterovirus (EV); Parechovirus (PeV);Chlamydia pneumoniae; Mycoplasma pneumoniae and Haemophilus influenzae type b (Hib); as per the manufacturer’s instruction. The reaction volume for each test was 25 μL, made up of 10 μL of nucleic acid and 15 μL of buffer/enzyme mix (Fast track mastermix). Amplification was performed in the ABI 7500 real-time PCR system thermocycler (Applied Biosystems, USA) using the following cycling conditions: 15 min at 42 °C, 3 min at 94 °C and 40 cycles of 8 s at 94 °C and 34 s at 60 °C. A fluorescence reading was taken at the 60 °C/34 s step in each cycle, and threshold cycle (Ct) values were determined by manual adjustment. Each sample was amplified in six parallel reactions, which contained primers and probes for four different targets, detecting viruses and bacteria, in addition to the internal control of the reaction. The positive and negative virus plasmid controls provided in the kit were included in all runs to monitor assay performance.
No follow-up or further contacts with the infants were kept post collection of nasopharyngeal swabs.
All data was entered in Microsoft excel sheet and frequencies of the etiological agent determined.
Results
One hundred infants (65 male infants, M:F = 1.86:1; Median age 6 mo; IQR: 5.5) with ALRTI admitted to the hospital during the study period (July 2014 through December 2015) were enrolled in the study. Fifty-two of them were diagnosed with bronchiolitis, while 48 were diagnosed with bronchopneumonia or pneumonia (radiographically, pulmonary infiltrates: 39, consolidation: 36 and hyperluscency: 25). As shown in Table 2, all the enrolled children had cough, fever and breathlessness with varying durations.
Eleven children had congenital heart disease, 10 were born prematurely while 27 had low birth-weight. Sixty-one infants were breastfed while 27 had history of exposure to smoke in their environment.
As shown in Table 3, 82 samples were positive for at least one pathogen. Of the positive samples, the common pathogens detected were Respiratory syncytial virus [RSV (29, 35.4%)], HRV (21, 25.6%) and Adenoviruses [ADV (18, 22%)]. Influenza A other than H1N1, Influenza B, Human coronavirus (hCoV 229E) and Enterovirus (EV) were not detected.
M. pneumoniae, C. pneumoniae and H. influenzae type b were the three bacteria detected.
As shown in Table 3, 28 (34%) were positive for multiple pathogens, while 54 (66%) were positive for a single pathogen. Among the single detections, RSV was the most common virus detected, while HRV was the most common virus detected among the multiple detections. hCoV NL 63, hCoV HKU1, PIV 1, 2, 4, PeV, M. pneumoniae and H. influenzae type b were detected only in co- detections andnever as single pathogens, while C. pneumoniae was detected only as a single pathogen. No co-detections involved more than three pathogens. (Table 4).
Three infants (aged 4–6 mo) with LRTI (severe bronchopneumonia) succumbed after developing progressive worsening in clinical condition. Two of these infants had evidence of congenital heart disease. The infant without congenital heart disease, who succumbed to his illness had several risk factors such as premature birth, low birth weight, malnutrition and absence of exclusive breast feeding.
Discussion
The burden of viral LRTIs among infants in western India is poorly understood, due to paucity of published reports. This communication, which has used the latest technological advancement for identification of viral agents aims to fill this gap in the knowledge. The study has shown that up to 82% of samples of infants with symptoms of respiratory infection had positive viral agent identification. RSV, HRV, ADV, HBoV and HMPV were the predominant viruses identified.
RSV (A/B) was detected in 35.4% of study samples and this figure is well within the proportions (10–58%) reported in earlier studies [6,7,8, 15, 16]. This suggests that maternal antibodies are ineffective in preventing RSV infections during infancy, leading to substantial morbidity. HRV, the second most common virus that was detected was prevalent in 25.6% of infants, which is within the range of 0.5–33% [1, 7, 15, 17] as reported by other investigators. Earlier considered as the most common cause of upper respiratory tract infection (URTI), and less commonly less a cause of LRTI, HRV has further established its role also as a common cause of LRTIs among infants [15]. ADV accounted for 22% of virus-detection-positive samples from infants. It has been described as one of the major causes of LRTI in children aged over 2 y and in adults, but not in infants [18]. The present study findings seem to suggest that a rethink on these views could be in order. Human bocavirus was found in 9.8% of infants with positive samples. This virus has been frequently identified in infants, although its role in disease pathogenesis is not yet clear. Longtin et al. documented that prevalence rates are higher in children under 2 y of age, and that they decrease with age, which implies that the protection due to antibodies against hBoV is acquired during early life [19]. The present study also detected the novel Influenza A (H1N1) pdm 09 virus in 6.1% (5/80) of the infants. Influenza A (other than the novel H1N1 pdm 09), was not detected, perhaps because the endemic Influenza A virus has been replaced or suppressed by the circulating novel H1N1 pdm 09 virus, as a result of the outbreak of H1N1virus that was reported starting early in 2015. The prevalence of H. influenzae type b (Hib) in developed countries has decreased, due to administration of Hib containing pentavalent vaccine; this pentavalent vaccine is included as part of the universal immunization program in several Indian states (Tamil Nadu, Kerala, Gujarat, Haryana, Karnataka, Goa, Jammu and Kashmir and Puducherry), but it was not implemented in Maharashtra during the period of this study, which explains detection of Hib in present study [20].
Earlier studies have mentioned a co- infection rate of 2–82% [1, 6,7,8, 21]. In this study, multiple pathogens were noted in 34% (28/82) of the positive samples. HRV was the commonest virus to be detected in multiple detections. Co-infections have been increasingly reported in infants [21]. The clinical significance of co-infections has not been fully elucidated. The authors cannot rule out the possibility that the high sensitivity of multiplex PCR may have led to the detection of residual nucleic acids from prior viral infections [1]. For viruses like ADV and hBoV, lymphoid tissues are known to serve as reservoirs. This may lead to their prolonged shedding in an asymptomatic individual, and cause transmission of these viruses to other susceptible individuals [22]. It is well-known that a few viruses like hCoV-NL63, HRV and hBoV continue to shed for a very long time after the onset of symptoms [23]. In addition, the duration of shedding of viruses may depend on whether it was the sole pathogen or was accompanied by other co-pathogens [24]. It is not surprising that HRV, ADV, hCoV NL63, hBoV were frequently detected in co-infections in present study. Because hBoV occurs so frequently with other viruses, it is still uncertain whether hBov is a pathogen, or just a concomitant bystander virus [19]. Similarly, due to nasal carriage of pathogenic bacteria like M. pneumoniae and Hib, the results of nasopharyngeal swab may be misleading. However, HRV cannot be neglected as a persister, since it has been shown to be associated with worsening outcome, which could be attributed to its ability to reduce the capacity of bronchial epithelial cells to proliferate and self-repair [25]. Thus, a positive result must be interpreted cautiously and correlated with the patient’s clinical symptoms.
The difference in the positivity rates between the above-mentioned studies for detecting viruses/bacteria and the present study could be explained by various factors, like the different age groups of the patients, the difference in the types of samples collected, different diagnostic methods used, and also wide inter-regional variations with different climates. The issue of association between presence of risk factors or multiple pathogens and mortality could not be determined due to the small sample available. It could be the object of future research.
Use of a sensitive and novel method to detect viral agents is the strength of the study. The specimen of choice for the detection of respiratory viruses is a nasopharyngeal aspirate (NPA) or a nasal wash [10, 14]. But both have their own disadvantages such as unpleasantness of the procedure, need for a suction device in the case of NPA, and the attendant risk of aspiration and generation of aerosol during the collection of nasal wash. Hence, these procedures are not feasible for widespread use in clinical practice. In contrast, collecting a nasopharyngeal swab is easy, painless, can be performed anywhere and does not require any additional device. Also, the, newly developed flocked swabs (which were used in the study) are more sensitive, and less traumatic for pediatric patients, and are also more acceptable to the parents [21]. The absence of a single test to detect all the etiological agents is the greatest hurdle in determining the etiological agent for cases with LRTI. Multiplex PCR has overcome this hurdle to some extent: it has the potential to both reduce the overall use of antibiotics and to improve the targeted use of antibiotics, which in turn can help control nosocomial transmission of viruses. However, the kit detects only the organisms mentioned above, and therefore may have missed other viruses and bacteria contributing to ALRTI. This may explain the 18 negative samples in the study. High concordance between the viruses identified using multiplex PCR on nasopharyngeal aspirates and those identified by studying pulmonary samples in patients with acute respiratory infections in previously published literature [26, 27] provides confidence that the organisms identified in this study in the nasopharyngeal aspirates were responsible for the lower respiratory infections. A case control study would be beneficial to know the presence of these viruses and bacteria in an asymptomatic infant. This could be an area of future research.
This study has established that it is feasible to use Multiplex PCR for detection of viral agents in the nasopharyngeal swabs. It has also provided a baseline data about the likely viruses that are detected in nasopharynx in infants with clinical manifestations of LRTI. Studies with larger sample with recruitment spanning all seasons can provide greater clarity about the contribution of various etiological agents.
References
Martins Júnior R, Carney S, Goldemberg D, et al. Detection of respiratory viruses by real-time polymerase chain reaction in outpatients with acute respiratory infection. Mem Inst Oswaldo Cruz. 2014;109:716–21.
Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2095–128.
Liu L, Johnson H, Cousens S, et al. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379:2151–61.
Yeolekar L, Damle R, Kamat A, Khude M, Simha V, Pandit A. Respiratory viruses in acute respiratory tract infections in Western India. Indian J Pediatr. 2008;75:341–5.
Kabra S, Lodha R, Broor S, Chaudhary R, Ghosh M, Maitreyi R. Etiology of acute lower respiratory tract infection. Indian J Pediatr. 2003;70:33–6.
Singh AK, Jain A, Jain B, et al. Viral aetiology of acute lower respiratory tract illness in hospitalized paediatric patients of a tertiary hospital: one-year prospective study. Indian J Med Microbiol. 2014;32:13–8.
Mathew JL, Singhi S, Ray P, et al. Etiology of community acquired pneumonia among children in India: prospective, cohort study. J Glob Health. 2015;5:050418.
Bharaj P, Sullender WM, Kabra SK, et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89.
Broor S, Parveen S, Bharaj P, et al. A prospective three-year cohort study of the epidemiology and virology of acute respiratory infections of children in rural India. PLoS One. 2007;2:e491.
Wright P, Cutt F. Generic protocol to examine the incidence of lower respiratory infection due to respiratory syncytial virus in children less than five years of age [Internet]. Geneva, Switzerland: World Health Organization, Department of Vaccines and Biologicals;2000. Available at:http://apps.who.int/iris/bitstream/handle/10665/66276/WHO_V_and_B_00.08_eng.pdf?sequence=1. Accessed 23 Aug 2018.
Centre for Disease Control & Prevention, World Food Programme. A Manual: Measuring and Interpreting Malnutrition and Mortality. Rome: CDS & World Food Programme: C; 2005: 17–20.
World Health Organization. WHO Child Growth Standards. Weight-for-age; girls birth to 2 years (z-scores) [Internet]. WHO; Available at: http://www.who.int/childgrowth/standards/cht_wfa_girls_z_0_2.pdf?ua=1. Accessed 5 Oct 2015.
World Health Organization. WHO Child Growth Standards. Weight for age; boys birth to 2 years (z scores). Available at: http://www.who.int/childgrowth/standards/cht_wfa_boys_z_0_2.pdf?ua=1. Accessed 5 Oct 2015.
Frayha H, Castriciano S, Mahony J, Chernesky M. Nasopharyngeal swabs and nasopharyngeal aspirates equally effective for the diagnosis of viral respiratory disease in hospitalized children. J Clin Microbiol. 1989;27:1387–9.
Hoffmann J, Rabezanahary H, Randriamarotia M, et al. Viral and atypical bacterial etiology of acute respiratory infections in children under 5 years old living in a rural tropical area of Madagascar. PLoS One. 2012;7:e43666.
Feng L, Li Z, Zhao S, et al. Viral etiologies of hospitalized acute lower respiratory infection patients in China, 2009-2013. PLoS One. 2014;9:e99419.
Espínola E, Russomando G, Aquino C, Basualdo W. Phylogeny-based classification of human rhinoviruses detected in hospitalized children with acute lower respiratory infection in Paraguay, 2010-2011. J Med Virol. 2013;85:1645–51.
Pretorius MA, Madhi SA, Cohen C, et al. Respiratory viral coinfections identified by a 10-plex real-time reverse-transcription polymerase chain reaction assay in patients hospitalized with severe acute respiratory illness--South Africa, 2009-2010. J Infect Dis. 2012;206:S159–65.
Longtin J, Bastien M, Gilca R, et al. Human bocavirus infections in hospitalized children and adults. Emerg Infect Dis. 2008;14:217–21.
Child Health Programme [Internet]. Minister of Health & Family Welfare; 2015 Available at: https://mohfw.gov.in/sites/default/files/54789632475632147555.pdf. Accessed 4 Oct 2015.
Meerhoff T, Houben M, Coenjaerts F, et al. Detection of multiple respiratory pathogens during primary respiratory infection: nasal swab versus nasopharyngeal aspirate using real-time polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 2010;29:365–71.
Proenca-Modena JL, Pereira Valera FC, Jacob M, et al. High rates of detection of respiratory viruses in tonsillar tissues from children with chronic adenotonsillar disease. PLoS One. 2012;7:e42136.
Nunes MC, Kuschner ZC, Rabede Z, et al. Clinical epidemiology of bocavirus, rhinovirus, two polyomaviruses and four coronaviruses in HIV-infected and HIV-uninfected South African children. PLoS One. 2014;9:e86448.
Martin E, Fairchok M, Stednick Z, Kuyperss J, Englund J. Epidemiology of multiple respiratory viruses in childcare attendees. J Infect Dis. 2013;207:982–9.
da Silva ER, Pitrez MCP, Arruda E, et al. Severe lower respiratory tract infection in infants and toddlers from a non-affluent population: viral etiology and co-detection as risk factors. BMC Infect Dis. 2013;13:41.
Robert S, Lhommet C, Brun C, et al. Diagnostic performance of multiplex PCR on pulmonary samples versus nasopharyngeal aspirates in community-acquired severe lower respiratory tract infections. J Clin Virol. 2018;108:1–5.
Zhang X, Lu A, Shi P, Wang L, Qian L. Diagnostic value of nasopharyngeal aspirates in children with lower respiratory tract infections. Chiness Med J. 2017;130:647–51.
Acknowledgements
The authors’ thank Puritan Medical Products, USA, for providing them flocked swabs with universal transport media (UT – 317).
Author information
Authors and Affiliations
Contributions
AAS: Assistance in conceptualization of the research project and developing protocol, conduct of research, data collection, interpretation of results, intellectual inputs in drafting the manuscript and approval of the final draft; JS: Conceptualized the research idea, supervised the conduct of the research, intellectual inputs in drafting the manuscript and approval of the final draft; SSB: Assistance in conceptualization of the research project and developing protocol, interpretation of results, intellectual inputs for improving the research paper and approval of the final draft.
Corresponding author
Ethics declarations
Conflict of Interest
None.
Source of Funding
Intramural research funds.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sonawane, A.A., Shastri, J. & Bavdekar, S.B. Respiratory Pathogens in Infants Diagnosed with Acute Lower Respiratory Tract Infection in a Tertiary Care Hospital of Western India Using Multiplex Real Time PCR. Indian J Pediatr 86, 433–438 (2019). https://doi.org/10.1007/s12098-018-2840-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12098-018-2840-8