Abstract
Background
Acute lower respiratory tract infections (ALRI) are the major cause of morbidity and mortality in young children worldwide. Information on viral etiology in ALRI from India is limited. The aim of the present study was to develop a simple, sensitive, specific and cost effective multiplex PCR (mPCR) assay without post PCR hybridization or nested PCR steps for the detection of respiratory syncytial virus (RSV), influenza viruses, parainfluenza viruses (PIV1–3) and human metapneumovirus (hMPV). Nasopharyngeal aspirates (NPAs) were collected from children with ALRI ≤ 5 years of age. The sensitivity and specificity of mPCR was compared to virus isolation by centrifugation enhanced culture (CEC) followed by indirect immunofluorescence (IIF).
Results
From April 2005–March 2007, 301 NPAs were collected from children attending the outpatient department or admitted to the ward of All India Institute of Medical Sciences hospital at New Delhi, India. Multiplex PCR detected respiratory viruses in 106 (35.2%) of 301 samples with 130 viruses of which RSV was detected in 61, PIV3 in 22, PIV2 in 17, hMPV in 11, PIV1 in 10 and influenza A in 9 children. CEC-IIF detected 79 viruses only. The sensitivity of mPCR was 0.1TCID50 for RSV and influenza A and 1TCID50 for hMPV, PIV1, PIV2, PIV3 and Influenza B. Mixed infections were detected in 18.8% of the children with viral infections, none detected by CEC-IIF. Bronchiolitis was significantly associated with both total viral infections and RSV infection (p < 0.05). History of ARI in family predisposed children to acquire viral infection (p > 0.05).
Conclusion
Multiplex PCR offers a rapid, sensitive and reasonably priced diagnostic method for common respiratory viruses.
Similar content being viewed by others
Background
Acute respiratory tract infections (ARI) are a leading cause of morbidity and mortality in children worldwide [1] accounting for about 30% of all childhood deaths in developing world [2]. Viruses account for 50–90% of acute lower respiratory tract infections (ALRI) in young children [3] with respiratory syncytial virus (RSV), parainfluenza viruses (PIV), influenza viruses A and B and human metapneumoviruses (hMPV) being most commonly identified [4–6].
Respiratory infections caused by above said viruses usually present with clinical features that are nearly indistinguishable [7]. The increased sensitivity of polymerase chain reaction (PCR) over conventional methods for the diagnosis of respiratory viral infections has been established previously [8]. However, organism-specific RT-PCR assays which require separate amplification of each virus under investigation are resource intensive, time consuming and labor intensive [9].
Multiplex PCRs (mPCR) detect multiple organisms in a single assay and are available either as commercial assays [9–12] or in-house assays [4, 5, 13–17]. Majority of the in-house mPCR assays have not included recently discovered respiratory pathogens and require validation of results by post PCR hybridization or semi/nested PCR which make the assay cost ineffective and increases chances of cross contamination. Commercially available mPCR assays are expensive and require dedicated instrumentation [18].
We developed a simplified and economical multiplex PCR assay without any post PCR hybridization/nested PCR steps for the detection of seven major respiratory viruses.
(This material was presented in part at the 7th Asia Pacific Congress of Medical Virology held at New Delhi, India in November 2006 and Options for the Control of Influenza VI held at Toronto, Canada in June 2007.)
Results
The primer set designed for PIV1 failed to amplify PIV1 RNA after repeated attempts. The primer set published by Osiowy in 1998 [17] was found to amplify PIV1 N gene successfully.
Standardization of cDNA synthesis
Ten Units of the AMV-RT enzyme, 500 ng of random hexamer primer (PdN6), 500 μM dNTP concentration and 8 U of RNAsin were found to be optimal for 25 μl cDNA synthesis.
Standardization of multiplex PCR
All seven sets of primers when combined led to mispairing and nonspecific amplification. After trying different combinations, it was observed that RSV, Influenza A and B viruses in one set and PIV1–3 and hMPV in another set gave specific amplification for each virus (Figure 1).
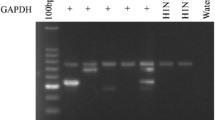
Standardization of two tube multiplex PCR for RSV, Influenza A&B viruses in first tube and PIV1–3 and hMPV in the second tube. Lane 1: Marker Ø X174 (Hae III digested). Lane 2: Amplicon forRSV showing band of 683 bp, Influenza A of 105 bp, Influenza B of 503 bp. Lane 3: Marker 100 bp ladder. Lane 4: Amplicon for PIV1 showing band of 84 bp, PIV2 of 197 bp, PIV3 of 266 bp, and hMPV of 440 bp.
Optimized reagent and PCR cycling conditions for first and second tube multiplex PCR
The optimized reagent concentrations for each tube were 25 pM of each primer, 400 μM of dNTPs, 2 mM MgCl2 and 6 U of Taq polymerase enzyme. The optimized cycling conditions for both tubes were: 94°C for 3 min followed by 35 cycles of 94°C for 1 min, 55°C for 1 min (53°C for 1 min for second tube) and 72°C for 1 min. Final extension was done at 72°C for 10 min for first tube and 7 min for second set.
Sensitivity and specificity of multiplex PCR
The sensitivity of detection by two tube multiplex PCR was 0.1TCID50 for RSV, Influenza A and 1TCID50 for hMPV, PIV1, PIV2, PIV3 and Influenza B. There were no non-specific amplification products against RNA from heterologous sources.
Detection of seven respiratory viruses in clinical samples
Study group
Three hundred and one children from OPD and ward with ALRI were enrolled in the study. Of the 166 children seen in the outpatient department, 137 (82.5%) had ALRI, 29 (17.5%) had severe ALRI and none had very severe ALRI (as per WHO classification). Of the 135 children admitted to pediatric ward, 35 (26%) had ALRI, 92 (68%) had severe ALRI and 8 (6%) had very severe ALRI (Table 1). More number of children with ALRI were seen in the OPD as compared to pediatric ward (p < 0.05, Pearson Chi square test). However, for severe ALRI, more children were admitted than seen in OPD (p < 0.05, Pearson Chi square test).
All the 301 children enrolled in the study were in the age range of 1–72 months with the median age of 11 months. The mean of their age was 15.6 ± 14 months. Among them 217 were males and 84 were females (Male: Female ratio = 2.6:1). It was observed that there was no significant difference between the age range of children with ALRI or severe ALRI from OPD or Ward (p > 0.05).
Detection of seven respiratory viruses in children with ALRI
Of the 301, 106 children (35.2%) had viral infections and were positive for 130 respiratory viruses. Of these 106 children with ALRI, 64 presented to the OPD and 42 were admitted to the ward. Of the 64 children who presented to OPD, 52 had ALRI and 12 had severe ALRI. Of the 42 children who were admitted, 8 had ALRI, 31 had severe ALRI and 3 had very severe ALRI (Table 2).
In 106 children in whom respiratory viruses were detected, the age range was 1–72 months with a median of 12 months and the mean age was 15.8 ± 13.8 months. In the PCR negative group, the age range was 1–61 months with a median of 10 months and the mean age was 15.5 ± 14.1 months. This difference was not statistically significant between the two groups. There was no significant difference in the male female ratio between the two groups.
RSV was detected in 61, PIV3 in 22, PIV2 in 17, hMPV in 11, PIV1 in 10 and Influenza A in 9 children respectively (Table 3). Figure 2 shows detection of single and mixed infections in some samples on which two tube multiplex PCR was applied. Of these, 86 were single virus infections and mixed infections were seen in 20 children (18.8% of the 106 children). Nested PCR for RSV identified the presence of RSV B in all 61 samples. Of the single infections, RSV comprised 50, hMPV 9, Influenza A and PIV3 8 each, PIV2 6 and PIV1 5.
Application of two tube multiplex PCR on clinical samples. Panel A. Lane 1: 100 bp DNA ladder. Lane 2: Clinical sample showing amplicon of 105 bp for FLU A. Lane 3: Clinical sample showing amplicon of 683 bp for RSV. Lane 4: Clinical sample showing amplicon of 440 bp for hMPV. Lane 5: Clinical sample showing amplicon of 266 bp for PIV3. Lane 3: Negative clinical sample. Panel B. Lane 1: 100 bp DNA adder. Lane 2: Clinical sample showing mixed infection of PIV1 (84 bp), PIV2 (197 bp) and PIV3 (266 bp).
It was seen that the percentage of virus detections by multiplex PCR were higher in the children with ALRI seen as outpatients (37.2%) as compared to those admitted to the ward (22.8%). Similarly, in the children with severe ALRI, seen as outpatients were higher percentage was positive for viruses (45%) as compared to those admitted to the ward (33.7%). It was observed that of all the mixed infections, 5.8% of them had ALRI whereas 7.8% of them had severe and very severe ALRI. Although severe ALRI was seen in higher number of children with mixed infections as compared to those with ALRI with mixed infections, the difference between the groups was not statistically significant.
Co-relation between multiplex PCR and tissue culture
The "gold standard" isolation in tissue culture by CEC-IIF detected 79 (61%) viruses as compared to 130 by multiplex PCR. CEC-IIF could not detect the presence of viruses in samples with mixed infections (data not shown). The sensitivity, specificity and likelihood ratio between the two assays is shown in Table 4.
Temporal distribution of respiratory viruses
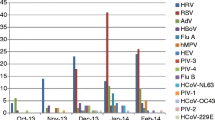
The number of RSV infections increased during late fall and peaked between October and January during the first year of the study. During the next year of the study, the distribution of RSV was scattered. PIVs were detected during the first year of the study, influenza A in winter months and hMPV in spring season (Figure 3).
Cost effectiveness of the multiplex PCR assay
The cost per sample detected by two tube multiplex PCR assay was USD16 (RNA extraction USD6, cDNA synthesis USD2.5 and two tube multiplex PCR USD4.5, equipment and personnel cost USD3) as compared to the cost per sample by culture being USD24 (Sample collection USD2, sample processing USD2, inoculation of sample on to 3 different cell lines USD8, indirect immunofluorescence USD3, visualization under fluorescent microscope USD3, equipment and personnel cost USD6).
Clinical symptoms
The clinical features, demographics and risk factors of children with viral infections and RSV alone were compared with the virus negative group (Table 5, 6). It was observed that significantly higher number of children below 12 months of age had RSV infection. Children presenting with preceding bronchiolitis were significantly associated with total viral infections and RSV infection (p < 0.05). Runny nose was significantly present in children with RSV infection (p < 0.05). Among risk factors, ARI in family was found to be associated with virus positive children (p < 0.005).
Discussion
The development of multiplex PCR for the detection of respiratory viruses as a rapid, sensitive and time saving technique has not gained priority in India even though ~0.5 million children die each year in this country due to ALRI each year, accounting for one fourth of the 1.9 million global ALRI deaths [19–21]. Among all the major ALRI causing viruses namely RSV, PIVs and influenza viruses A and B, the presence of RSV has been documented to be the most commonly identified pathogen followed by PIV3 [22]. In the present study, we standardized and evaluated an economical two-tube multiplex PCR assay devoid of any further confirmatory steps. The present assay reagents costs were mere USD16/reaction in contrast to USD90/reaction [18] reported for a commercial assay. The sensitivity of our multiplex PCR assay was similar or better than previously described mPCR assays for these viruses [5, 16, 17, 23, 24]. We did not make direct comparisons of the performance of the different assays in our laboratory.
In the present study we could culture majority of the viruses detected by mPCR with the exception of RSV which is known to be highly thermolabile [25]. The detection rate of viruses was similar to detection rate reported earlier [16, 17, 24, 26]. It was observed that a higher proportion of virus positive children presented to the OPD than the ward, similar to a study from Taiwan [26] and could be due to the fact that the patients present earlier after onset of symptoms to the OPD as compared to getting admitted to the Ward. However, this could also be because severe disease is more likely to be admitted to the hospital and caused by bacteria than virus [27]. A higher proportion of males were found to have infection with respiratory viruses as compared to females as reported earlier [28, 29].
RSV was most commonly identified viral pathogen similar to previously described viral identifications by mPCR [16, 17, 28, 29]. PIVs were the second most frequently identified pathogens [29] followed by hMPV [22, 28–30] and Influenza A virus infections [16, 24, 28].
The detection rate of co-infections was similar to previously reported multiplex PCR studies [5, 14–17, 28, 29]. It was observed that higher percentage of children with mixed infections had severe and very severe ALRI as compared to ALRI. Previous studies have shown that co-infection with different respiratory viruses might lead to a more severe disease [31] or multiple viruses have been detected from patients with severe disease [32].
ALRI caused by RSV was more common in younger children as reported previously [28]. RSV and hMPV were associated with bronchiolitis [28, 29, 33, 34]. PIVs and Influenza viruses were associated with pneumonia similar to previous findings [28, 29]. However, the number of all the viral detections except RSV was too few to comment on the association of the virus with bronchiolitis or pneumonia.
In the present study, RSV was detected during the fall season similar to previously described studies from our laboratory [35–37]. The rest of the virus identifications were few and their seasonality cannot be commented upon.
Conclusion
In conclusion, we report a simplified multiplex PCR for the detection of seven respiratory viruses in samples from children with ALRI. This assay was found to be more sensitive, less time consuming and economical than virus isolation. Multiplex PCR format allowed the detection of co-infections which cannot be done using monoplex PCR or culture as shown in the present study.
Methods
Patient Specimens
Between April 2005 and March 2007, nasopharyngeal aspirates (NPAs) were collected from children ≤ 5 years of age with ALRI, severe ALRI and very severe ALRI as per WHO criteria [38] and are shown in Table 7.
The children were either seen at the Outpatient Department (OPD) or admitted to the Pediatric Ward of All India Institute of Medical Sciences (AIIMS) Hospital, New Delhi, India. The demographic profile of child, clinical symptoms and risk factors were recorded in a predesigned proforma. NPAs were collected and processed as described earlier [39].
Standard strains of viruses
Standard strains of 9 viruses namely human respiratory syncytial virus (A2 and 18537), PIV1 (Washington/1964), PIV2 (Greer), PIV3 (D10025), influenza A {H1N1 (A/New Caledonia/20/99) and H3N2 (A/Panama/2007/99)} and B viruses and human metapneumovirus hMPV (Can 97–83) were cultured in Hep-2, MDCK and LLCMK-2 cells as described elsewhere [39–41].
RNA extraction
RNA was extracted from standard strain of the virus by guanidinium thiocyanate method [42] and 500 μl of NPA using RNeasy kit (Qiagen, GmBH, Germany) described previously [39].
cDNA synthesis
cDNA synthesis was optimized using 5–20 units of AMV-RT enzyme, 500 ng -1000 ng random hexamer primer (PdN6), 0.1–2 mM dNTPs, 4–8 units of RNAsin (all reagents from Promega, Madison, WI, USA) and 5–10 μl of RNA in a 25 μl reaction volume.
Primer Designing
For RSV, PIVs and hMPV primers were designed from nucleocapsid region and for Influenza (A and B) from the matrix region using sequences available in GenBank, using program OLIGO (Molecular Biology Insights, Cascade, CO, USA, http://oligo.net) and oligonucleotide T m calculator (http://idtdna.com). The sequence of all the seven sets of primers and nested primers for RSV group A and B are shown in Table 8.
PCR standardization
cDNA was synthesized from pooled RNA of different viruses to generate template for multiplex PCR. Parameters that were optimized included different concentrations of primers, dNTPs, magnesium chloride (MgCl2), Taq polymerase, adjuvants (DMSO and glycerol) and cycling conditions for a 25 μl reaction. If RSV was detected then nested PCR was done for typing of RSV into group A or B. All the PCR reactions were conventional block PCR assays, carried out in GeneAmp® PCR System 9700 (Applied Biosystems, USA) using plasticware from Axygen Scientific, USA.
An internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included to check the presence of inhibitors of the RT-PCR assay.
Sensitivity and specificity of the Multiplex PCR
The sensitivity of the multiplex PCR assay was determined by TCID50 using Reed and Muench method [43]. Inter and intra assay specificity of the primers was tested with RNA extracted from RSV A and B, PIV1–3, Influenza A and B viruses, hMPV, enteroviruses, cytomegalovirus, herpes simplex virus 1 & 2.
Virus isolation by centrifugation enhanced culture
Virus isolations were done using centrifugation enhanced culture (CEC) followed by indirect immunofluorescence (IIF) as described previously [35].
Costing methods
Costs are reported in this manuscript using United States dollar values, with 2006 taken as the reference year for reporting unit prices.
Metrological data
The environmental factors namely rainfall (cm), temperature (°C) and humidity (RH in %) were acquired from the India Meteorological Department, Regional Meteorological Centre, New Delhi, India.
Statistical analysis
Statistical analysis was carried out using STATA 9.0 (College station, Texas, USA). Data were presented as number or median (Range). Validity of multiplex PCR in comparison to CEC-IIF was assessed using sensitivity (95% CI), specificity (95% CI) and likelihood ratio. The association between clinical features at the time of presentation and virus detection was tested using Chi-square/Fisher's exact test as appropriate and OR (95% CI) was also calculated. A p value of < 0.05 was considered statistically significant.
Authors' information
Preeti Bharaj is a PhD scholar from Department of Microbiology, All India Institute of Medical Sciences, Ansari Nagar, New Delhi, 110029, India.
References
Hijazi Z, Pacsa A, Eisa S, el Shazli A, abd el-Salam RA: Laboratory diagnosis of acute lower respiratory tract viral infections in children. J Trop Pediatr 1996,42(5):276-80. 10.1093/tropej/42.5.276
Hinman AR: Global progress in infectious disease control. Vaccine 1998, 16: 1116-21. 10.1016/S0264-410X(98)80107-2
Glezen WP, Loda FA, Clyde WA Jr, Senior RJ, Sheaffer CI, Conley WG, Denny FW: Epidemiologic patterns of acute lower respiratory disease of children in a pediatric group practice. J Pediatr 1971,78(3):397-406. 10.1016/S0022-3476(71)80218-4
Fan J, Henrickson KJ, Savatski LL: Rapid simultaneous diagnosis of infections with respiratory syncytial viruses A and B, influenza viruses A and B, and human parainfluenza virus types 1, 2, and 3 by multiplex quantitative reverse transcription-polymerase chain reaction-enzyme hybridization assay (Hexaplex). Clin Infect Dis 1998,26(6):1397-1402. 10.1086/516357
Bellau-Pujol S, Vabret A, Legrand L, Dina J, Gouarin S, Petitjean-Lecherbonnier J, Pozetto B, Ginevra C, Freymuth F: Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods 2005,126(1–2):53-63. 10.1016/j.jviromet.2005.01.020
Broor S, Bharaj P: Avian and human metapneumovirus. Ann N Y Acad Sci 2007, 1102: 66-85. 10.1196/annals.1408.005
Debbia EA, Schito GC, Zoratti A, Gualco L, Tonoli E, Marchese A: Epidemiology of major respiratory pathogens. J Chemother 2001,1(1):205-10.
Weinberg GA, Erdman DD, Edwards KM, Hall CB, Walker FJ, Griffin MR, Schwartz B, New Vaccine Surveillance Network Study Group: Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J Infect Dis 2004,189(4):706-10. 10.1086/381456
Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC: Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol 2004,42(4):1564-9. 10.1128/JCM.42.4.1564-1569.2004
Fan J, Henrickson KJ: Rapid diagnosis of human parainfluenza virus type 1 infection by quantitative reverse transcription-PCR-enzyme hybridization assay. J Clin Microbiol 1996, 34: 1914-17.
Nolte FS, Marshall DJ, Rasberry C, Schievelbein S, Banks GG, Storch GA, Arens MQ, Buller RS, Prudent JR: MultiCode-PLx System for Multiplexed Detection of Seventeen Respiratory Viruses. J Clin Microbiol 2007,45(9):2779-86. 10.1128/JCM.00669-07
Marshall DJ, Reisdorf E, Harms G, Beaty E, Moser MJ, Lee WM, Gern JE, Nolte FS, Shult P, Prudent JR: Evaluation of a Multiplexed PCR Assay for Detection of Respiratory Viral Pathogens in a Public Health Laboratory Setting Prudent. J Clin Microbiol 2007,45(12):3875-82. 10.1128/JCM.00838-07
Aguilar JC, Perez-Brena MP, Garcia ML, Cruz N, Erdman DD, Echevarria JE: Detection and identification of human parainfluenza viruses 1, 2, 3, and 4 in clinical samples of pediatric patients by multiplex reverse transcription-PCR. J Clin Microbiol 2000,38(3):1191-5.
Coiras MT, Perez-Brena P, Garcia ML, Casas I: Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested-PCR assay. J Med Virol 2003,69(1):132-44. 10.1002/jmv.10255
Coiras MT, Aguilar JC, Garcia ML, Casas I, Perez-Brena P: Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J Med Virol 2004,72(3):484-95. 10.1002/jmv.20008
Grondahl B, Puppe W, Hoppe A, Kuhne I, Weigl JA, Schmitt HJ: Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J Clin Microbiol 1999,37(1):1-7.
Osiowy C: Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J Clin Microbiol 1998,36(11):3149-54.
Hindiyeh M, Hillyard DR, Carroll KC: Evaluation of the Prodesse Hexaplex multiplex PCR assay for direct detection of seven respiratory viruses in clinical specimens. Am J Clin Pathol 2001,116(2):218-24. 10.1309/F1R7-XD6T-RN09-1U6L
Ahmad OB, Lopez AD, Inoue M: The decline in child mortality: a reappraisal. Bull World Health Organ 2000,78(10):1175-91.
Reddaiah VP, Kapoor SK: Acute respiratory infections in rural underfives. Indian J Pediatr 1988,55(3):424-426. 10.1007/BF02810367
Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C: Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis 2002,2(1):25-32. 10.1016/S1473-3099(01)00170-0
Hall CB: Respiratory syncytial virus and parainfluenza virus. N Engl J Med 2001,21;344(25):1917-28. 10.1056/NEJM200106213442507
Puppe W, Weigl JA, Aron G, Grondahl B, Schmitt HJ, Niesters HG, Groen J: Evaluation of a multiplex reverse transcriptase PCR ELISA for the detection of nine respiratory tract pathogens. J Clin Virol 2004,30(2):165-74. 10.1016/j.jcv.2003.10.003
Syrmis MW, Whiley DM, Thomas M, Mackay IM, Williamson J, Siebert DJ, Nissen MD, Sloots TP: A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn 2004,6(2):125-31.
Falsey AR, Walsh EE: Respiratory syncytial virus infection in adults. Clin Microbiol Rev 2000,3(3):371-84. 10.1128/CMR.13.3.371-384.2000
Tsai HP, Kuo PH, Liu CC, Wang JR: Respiratory Viral Infections among Pediatric Inpatients and Outpatients in Taiwan from 1997 to 1999. J Clin Microbiol 2001,39(1):111-8. 10.1128/JCM.39.1.111-118.2001
Huq F, Rahman M, Nahar N, Alam A, Haque M, Sack DA, Butler T, Haider R: Acute lower respiratory tract infection due to virus among hospitalized children in Dhaka, Bangladesh. Rev Infect Dis 1990, 12 Suppl 8: S982-S987.
Choi EH, Lee HJ, Kim SJ, Eun BW, Kim NH, Lee JA, Lee JH, Song EK, Kim SH, Park JY, Sung JY: The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000–2005. Clin Infect Dis 2006, 43: 585-92. 10.1086/506350
Thomazelli LM, Vieira S, Leal AL, Sousa TS, Oliveira DB, Golono MA, Gillio AE, Stwien KE, Erdman DD, Durigon EL: Surveillance of eight respiratory viruses in clinical samples of pediatric patients in southeast Brazil. J Pediatr (Rio J) 2007,83(5):422-8. 10.1590/S0021-75572007000600005
Banerjee S, Bharaj P, Sullender W, Kabra SK, Broor S: Human metapneumovirus infections among children with acute respiratory infections seen in a large referral hospital in India. J Clin Virol 2007,38(1):70-2. 10.1016/j.jcv.2006.07.003
Kaida A, Kubo H, Goto K, Shiomi M, Kohdera U, Iritani N: Co-infection of human metapneumovirus with adenovirus or respiratory syncytial virus among children in Japan. Microbiol Immunol 2007,51(7):679-83.
McNamara PS, Flanagan BF, Smyth RL, Hart CA: Impact of human metapneumovirus and respiratory syncytial virus co-infection in severe bronchiolitis. Pediatr Pulmonol 2007,42(8):740-3. 10.1002/ppul.20649
Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE Jr: Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med 2004,350(5):443-50. 10.1056/NEJMoa025472
Ebihara T, Endo R, Kikuta H, Ishiguro N, Ishiko H, Hara M, Takahashi Y, Kobayashi K: Human metapneumovirus infection in Japanese children. J Clin Microbiol 2004, 42: 126-32. 10.1128/JCM.42.1.126-132.2004
Maitreyi RS, Broor S, Kabra SK, Ghosh M, Seth P, Dar L, Prasad AK: Rapid detection of respiratory viruses by centrifugation enhanced cultures from children with acute lower respiratory tract infections. J Clin Virol 2000,16(1):41-7. 10.1016/S1386-6532(99)00075-X
Broor S, Parveen S, Bharaj P, Prasad VS, Srinivasulu KN, Sumanth KM, Kapoor SK, Fowler K, Sullender WM: A prospective three-year cohort study of the epidemiology and virology of acute respiratory infections of children in rural India. PLoS ONE 2007,2(6):e491. 10.1371/journal.pone.0000491
Yusuf S, Piedimonte G, Auais A, Demmler G, Krishnan S, Van Caeseele P, Singleton R, Broor S, Parveen S, Avendano L, Parra J, Chavez-Bueno S, Murguía De Sierra T, Simoes EA, Shaha S, Welliver R: The relationship of meteorological conditions to the epidemic activity of respiratory syncytial virus. Epidemiol Infect 2007,135(7):1077-90. 10.1017/S095026880600776X
Integrated management of childhood illness In World Health Organization: WHO/FCH/CAH/00.12. HANDBOOK. WHO; 2000.
Parveen S, Sullender WM, Fowler K, Lefkowitz EJ, Kapoor SK, Shobha Broor S: Genetic Variability in the G Protein Gene of Group A and B Respiratory Syncytial Viruses from India. J Clin Microbiol 2006,44(9):3055-64. 10.1128/JCM.00187-06
Tao T, Durbin AP, Whitehead SS, Davoodi F, Collins PL, Murphy BR: Recovery of a fully viable chimeric human parainfluenza virus (PIV) type 3 in which the hemagglutinin-neuraminidase and fusion glycoproteins have been replaced by those of PIV type 1. J Virol 1998,72(4):2955-61.
Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD: A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 2001,7(6):719-24. 10.1038/89098
Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987,162(1):156-9. 10.1016/0003-2697(87)90021-2
Reed LJ, Muench H: A simple method of estimating fifty percent endpoints. The American Journal of Hygiene 1938, 27: 493-7.
Gueudin M, Vabret A, Petitjean J, Gouarin S, Brouard J, Freymuth F: Quantitation of respiratory syncytial virus RNA in nasal aspirates of children by real-time RT-PCR assay. J Virol Methods 2003,109(1):39-45. 10.1016/S0166-0934(03)00042-9
Acknowledgements
The funding for the research was supported by NIH Project No. 1 R21 AI59792-01.
We acknowledge the Indian Council of Medical Research (ICMR), India for supporting Preeti Bharaj via fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PB carried out all the molecular and culture based assays and prepared the manuscript. WMS contributed in analysis for the paper and drafting the manuscript. SKK, CJ and VT clinically analyzed the pediatric patients and collected samples from them. HSC, SK, LD helped in analyzing data, drafting and critical reviewing of the manuscript. SB conceived the idea, helped in analysis of the data, participated in its design and coordination and helped to frame the manuscript. All the authors have contributed to, seen and approved the final submitted version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bharaj, P., Sullender, W.M., Kabra, S.K. et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J 6, 89 (2009). https://doi.org/10.1186/1743-422X-6-89
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-6-89