Abstract
Introduction
Anthracyclines have various mechanisms of action that in the end lead to DNA double-strand breaks. Single-nucleotide polymorphisms (SNPs) in DNA repair genes may alter the protein function, affecting DNA repair proficiency and, therefore, the efficiency of DNA damaging chemotherapy. We have analysed whether SNPs in DNA repair genes (XRCC1, XRCC3 and XPD) could be useful to predict the response to anthracyclines in patients with early-stage breast cancer (EBC).
Methods
Peripheral blood samples from 150 patients with EBC were used for genotyping XRCC3Thr241Met, XRCC1Arg399Gln and XPDLys751Gln. Genotypes were correlated with survival outcomes.
Results
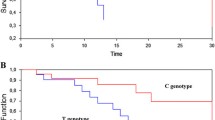
Eighty-four patients received treatment with chemotherapy regimens containing anthracyclines. In this group, patients with XRCC1Arg399Arg had a significant improvement in 5-year Disease Free Survival (DFS) compared with those with the Arg/Gln and Gln/Gln variants (84 vs 46 %, p = 0.026). In the multivariate analysis, XRCC1Arg399Arg was reported as an independent prognostic factor for DFS (HR 0.4, CI-95 % 0.2–0.9, p = 0.035). Patients with the XRCC3 Met241Met genotype presented better 5-year OS than those carrying the Thr/Thr and Met/Thr variants (100 vs 70 %, p = 0.030). A multivariate analysis for OS confirmed the independent prognostic value of XRCC3 Met241Met (HR 0.15, CI-95 % 0.02–0.90, p = 0.048). These differences were not significant when patients receiving other chemotherapy treatments, different from anthracyclines, were also considered (n = 150). XPDLys751Lys was associated with older age at diagnosis than the Lys/Gln and Gln/Gln genotypes (65 vs 58 years, p < 0.0001).
Conclusions
XRCC3Thr241Met and XRCC1Arg399Gln may be predictive of survival outcome in EBC patients treated with anthracycline-based chemotherapy regimens.


Similar content being viewed by others
References
Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18(3):581–92.
Autier P, Boniol M, La Vecchia C, Vatten L, Gavin A, Hery C, et al. Disparities in breast cancer mortality trends between 30 European countries: retrospective trend analysis of WHO mortality database. BMJ. 2010;341:c3620.
Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2010;62(4):220–41.
Di Marco A, Silvestrini R, Di Marco S, Dasdia T. Inhibiting effect of the new cytotoxic antibiotic daunomycin on nucleic acids and mitotic activity of HeLa cells. J Cell Biol. 1965;27(3):545–50.
Feinstein E, Canaani E, Weiner LM. Dependence of nucleic acid degradation on in situ free-radical production by Adriamycin. Biochemistry. 1993;32(48):13156–61.
Skladanowski A, Konopa J. Interstrand DNA crosslinking induced by anthracyclines in tumour cells. Biochem Pharmacol. 1994;47(12):2269–78.
Swift LP, Rephaeli A, Nudelman A, Phillips DR, Cutts SM. Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Res. 2006;66(9):4863–71.
Sinha BK, Mimnaugh EG, Rajagopalan S, Myers CE. Adriamycin activation and oxygen free radical formation in human breast tumor cells: protective role of glutathione peroxidase in Adriamycin resistance. Cancer Res. 1989;49(14):3844–8.
Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226(4673):466–8.
Zunino F, Gambetta R, Di Marco A, Zaccara A, Luoni G. A comparison of the effects of daunomycin and Adriamycin on various DNA polymerases. Cancer Res. 1975;35(3):754–60.
Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–74.
Munro AF, Cameron DA, Bartlett JM. Targeting anthracyclines in early breast cancer: new candidate predictive biomarkers emerge. Oncogene. 2010;29(38):5231–40.
Saffi J, Agnoletto MH, Guecheva TN, Batista LF, Carvalho H, Henriques JA, et al. Effect of the anti-neoplastic drug doxorubicin on XPD-mutated DNA repair-deficient human cells. DNA Repair (Amst). 2010;9(1):40–7.
Spencer DM, Bilardi RA, Koch TH, Post GC, Nafie JW, Kimura K, et al. DNA repair in response to anthracycline-DNA adducts: a role for both homologous recombination and nucleotide excision repair. Mutat Res. 2008;638(1–2):110–21.
Au WW, Salama SA, Sierra-Torres CH. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ Health Perspect. 2003;111(15):1843–50.
Vodicka P, Kumar R, Stetina R, Sanyal S, Soucek P, Haufroid V, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25(5):757–63.
Vodicka P, Stetina R, Polakova V, Tulupova E, Naccarati A, Vodickova L, et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis. 2007;28(3):657–64.
Synowiec E, Stefanska J, Morawiec Z, Blasiak J, Wozniak K. Association between DNA damage, DNA repair genes variability and clinical characteristics in breast cancer patients. Mutat Res. 2008;648(1–2):65–72.
Bewick MA, Conlon MS, Lafrenie RM. Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after treatment for metastatic breast cancer. J Clin Oncol. 2006;24(36):5645–51.
Han S, Zhang HT, Wang Z, Xie Y, Tang R, Mao Y, et al. DNA repair gene XRCC3 polymorphisms and cancer risk: a meta-analysis of 48 case-control studies. Eur J Hum Genet. 2006;14(10):1136–44.
Li H, Ha TC, Tai BC. XRCC1 gene polymorphisms and breast cancer risk in different populations: a meta-analysis. Breast. 2009;18(3):183–91.
Clarkson SG, Wood RD. Polymorphisms in the human XPD (ERCC2) gene, DNA repair capacity and cancer susceptibility: an appraisal. DNA Repair (Amst). 2005;4(10):1068–74.
Li C, Jiang Z, Liu X. XPD Lys(751)Gln and Asp (312)Asn polymorphisms and bladder cancer risk: a meta-analysis. Mol Biol Rep. 2010;37(1):301–9.
Ye W, Kumar R, Bacova G, Lagergren J, Hemminki K, Nyren O. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case-control study in Sweden. Carcinogenesis. 2006;27(9):1835–41.
Manuguerra M, Saletta F, Karagas MR, Berwick M, Veglia F, Vineis P, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol. 2006;164(4):297–302.
Sun H, Qiao Y, Zhang X, Xu L, Jia X, Sun D, et al. XRCC3 Thr241Met polymorphism with lung cancer and bladder cancer: a meta-analysis. Cancer Sci. 2010;101(8):1777–82.
Nowosielska A, Marinus MG. Cisplatin induces DNA double-strand break formation in Escherichia coli dam mutants. DNA Repair (Amst). 2005;4(7):773–81.
de las Penas R, Sanchez-Ronco M, Alberola V, Taron M, Camps C, Garcia-Carbonero R, et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17(4):668–75.
Krupa R, Synowiec E, Pawlowska E, Morawiec Z, Sobczuk A, Zadrozny M, et al. Polymorphism of the homologous recombination repair genes RAD51 and XRCC3 in breast cancer. Exp Mol Pathol. 2009;87(1):32–5.
Rodriguez AA, Makris A, Wu MF, Rimawi M, Froehlich A, Dave B, et al. DNA repair signature is associated with anthracycline response in triple negative breast cancer patients. Breast Cancer Res Treat. 2010;123(1):189–96.
Acknowledgments
ESMO Foundation awarded Elena Castro with an IMPAKT 2010 travel grant for the presentation of this work. We are grateful to Dr. Joo Ern Ang (Sutton, UK) for the editorial assistance provided.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Castro, E., Olmos, D., Garcia, A. et al. Role of XRCC3, XRCC1 and XPD single-nucleotide polymorphisms in survival outcomes following adjuvant chemotherapy in early stage breast cancer patients. Clin Transl Oncol 16, 158–165 (2014). https://doi.org/10.1007/s12094-013-1055-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-013-1055-8