Abstract
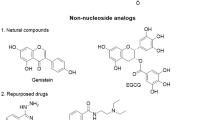
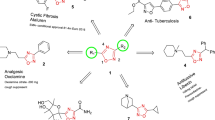
Multidrug resistance in Mycobacterium tuberculosis (M. Tb) and its coexistence with HIV are the biggest therapeutic challenges in anti-M. Tb drug discovery. The current study reports a Virtual Screening (VS) strategy to identify potential inhibitors of Mycobacterial cyclopropane synthase (CmaA1), an important M. Tb target considering the above challenges. Five ligand-based pharmacophore models were generated from 40 different conformations of the cofactors of CmaA1 taken from molecular dynamics (MD) simulations trajectories of CmaA1. The screening abilities of these models were validated by screening 23 inhibitors and 1398 non-inhibitors of CmaA1. A VS protocol was designed with four levels of screening i.e., ligand-based pharmacophore screening, structure-based pharmacophore screening, docking and absorption, distribution, metabolism, excretion and the toxicity (ADMET) filters. In an attempt towards repurposing the existing drugs to inhibit CmaA1, 6,429 drugs reported in DrugBank were considered for screening. To find compounds that inhibit multiple targets of M. Tb as well as HIV, we also chose 701 and 11,109 compounds showing activity below 1 μM range on M. Tb and HIV cell lines, respectively, collected from ChEMBL database. Thus, a total of 18,239 compounds were screened against CmaA1, and 12 compounds were identified as potential hits for CmaA1 at the end of the fourth step. Detailed analysis of the structures revealed these compounds to interact with key active site residues of CmaA1.

The paper presents generation of dynamic ligand-based pharmacophore model for Mycobacterial cyclopropane synthase and their validation. Aiming towards drug repositioning and selection of multi-target inhibitors, selected compounds from DrugBank and ChEMBL have been screened by a four-step virtual screening using the models and 12 potential compounds are identified.







Similar content being viewed by others
References
Balganesh T S, Alzari P M and Cole S T 2008 Trends Pharmacol. Sci. 29 576
WHO 2014 Global Tuberculosis Report. http//www.who.int/tb/en/ (accessed on 20/06/2015)
Zumla A, George A, Sharma V, Herbert N and Ilton B M 2013 Lancet 382 1765
Varghese G M, Janardhanan J, Ralph R and Abraham O C 2013 Curr. Infect. Dis. Rep. 15 77
Mdluli K, Kaneko T and Upton A 2015 Cold Spring Harb. Perspect. Med. doi:10.1101/cshperspect.a021154
Kandel D D, Raychaudhury C and Pal D 2014 J. Mol. Model. 20 2164
Raychaudhury C, Kandel D D and Pal D 2014 Croat. Chem. Acta 87 39
Lamichhane G 2011 Trends Mol. Med. 17 25
Choudhury C, Priyakumar U D and Sastry G N 2014 J. Struct. Biol. 187 38
Choudhury C, Priyakumar U D and Sastry G N J. Chem. Inf. Model. 55 848
Wishart D S, Knox C, Guo A C, Shrivastava S, Hassanali M, Stothard P, Chang Z and Woolsey J 2006 Nucleic Acids Res. 34 D668
Tobinick E L 2009 Drug News Perspect. 22 119
Bohari M H and Sastry G N 2012 J. Mol. Model. 18 4263
Bento A P, Gaulton A, Hersey A, Bellis L J, Chambers J, Davies M, Krüger F A, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R and Overington J P 2014 Nucleic Acids Res. 42 D1083
Kinnings S L, Liu N, Buchmeier N, Tonge P J, Xie L and Bourne P. 2009 PLoS Comput. Biol. 5 e1000423
Carlson H A, Masukawa K M, Rubins K, Bushman F D, Jorgensen W L, Lins R D, Briggs J M and McCammon J A 2000 J. Med. Chem. 43 2100
Meagher K L and Carlson H A 2004 J. Am. Chem. Soc. 126 13276
Meagher K L and Carlson H A 2005 Proteins Struct. Funct. Bioinf. 58 119
Damm K L and Carlson H A 2007 J. Am. Chem. Soc. 129 8225
Kitchen D B, Decornez H, Furr J R and Bajorath J 2004 Nat. Rev. Drug Discov. 3 935
Kubinyi H 1997 Drug Discov. Today 2 457
Srivastava H K and Sastry G N 2012 J. Chem. Inf. Model. 52 3088
Carlson H A 2002 Curr. Opin. Chem. Biol. 6 447
Srivastava H K, Choudhury C and Sastry G N 2012 Med. Chem. 8 811
Badrinarayan P and Sastry G N 2011 Comb. Chem. High Thr. Scr. 14 840
Badrinarayan P and Sastry G N 2012 J. Mol. Graph. Modell. 34 89
Badrinarayan P and Sastry G N 2014 PLoS One 9 e113773
Reddy A S, Pati S P, Kumar P P, Pradeep H N and Sastry G N 2007 Curr. Protein Pept. Sci. 8 329
Badrinarayan P and Sastry G N 2013 Curr. Pharm. Des. 19 4714
Selick H E, Beresford A P and Tarbit M H 2002 Drug Discov. Today 7 109
Kubinyi H 2003 Nat. Rev. Drug Discov. 2 665
de Waterbeemd H V and Gifford E 2003 Nat. Rev. Drug Discov. 2 192
Oprea T I, Davis A M, Teague S J and Leeson P D 2001 J. Chem. Inf. Comput. Sci. 41 1308
Schrödinger Release 2015-4: Maestro, version 10.4, 2015, Schrödinger, LLC, New York, NY
Dixon S L, Smondyrev A M, Knoll E H, Rao S N, Shaw D E and Friesner R A 2006 J. Comput. Aided. Mol. Des. 20 647
Anuradha A, Trivelli X, Guérardel Y, Dover L G, Besra G S, Sacchettini J C, Reynolds R C, Coxon G D and Kremer L 2007 PLoS One 12 e1343
LigPrep, version, 2.5 2012 Schrödinger, LLC, New York, NY
Glide, version, 5.8 2012, Schrödinger, LLC, New York, NY
Friesner R A, Murphy R B, Repasky M P, Frye L L, Greenwood J R, Halgren T A, Sanschagrin P C and Mainz D T 2006 J. Med. Chem. 49 6177
QikProp version 3.5 2012, Schrödinger, LLC, New York, NY
Yang S Y 2010 Drug Discov. Today 15 444
Saha S and Sastry G N 2015 J. Phys. Chem. B 119 11121
Mahadevi A S and Sastry G N 2013 Chem. Rev. 113 2100
Alhamadsheh M M, Waters N C, Sachdeva S, Lee P and Reynolds K A 2008 Bioorg. Med. Chem. Lett. 18 6402
Guüzel Ö, Maresca A, Scozzafava A, Salman A, Balaban A T and Supuran C T 2009 J. Med. Chem. 52 4063
Shu-Feng Z, Wang L, Di Y M, Xue C C, Duan W, Li C G and Li Y 2008 Curr. Med. Chem. 15 1981
Fardis M, Jin H, Jabri S, Cai R Z, Mish M, Tsiang M and Kim C U 2006 Bioorg. Med. Chem. Lett. 16 4031
Artico M, Santo R D, Costi R, Novellino E, Greco G, Massa S, Tramontano E, Marongiu M E, Montis A D and Colla P L 1998 J. Med. Chem. 41 3948
Maurin C, Lion C, Bailly F, Touati N, Vezin H, Mbemba G, Mouscadet J F, Debyser Z, Witvrouw M and Cotelle P 2010 Bioorg. Med. Chem. 18 5194
Porcari A R, Ptak R G, Borysko K Z, Breitenbach J M, Drach J C and Townsend L B 2000 J. Med. Chem. 43 2457
Acknowledgements
CC thanks Department of Science and Technology (DST), New Delhi for financial assistance through INSPIRE fellowship. UDP thanks DAE-BRNS for financial assistance. GNS thanks CSIR, New Delhi for financial support in the form of XII five year project (GENESIS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information (SI)
Scheme S1 showing the reference CmaA1 inhibitors, List S1 gives the details of the dataset download and preparation, table S1 giving the details (number of compounds matched, docking and fitness scores, etc.) of all the ligand-based pharmacophore models, table S2–S5 giving the docking scores and QuickProp Descriptors of all screened compounds and List S2 giving the descriptions of the Quickprop descriptors are provided at www.ias.ac.in/chemsci.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
CHOUDHURY, C., PRIYAKUMAR, U.D. & SASTRY, G.N. Dynamic ligand-based pharmacophore modeling and virtual screening to identify mycobacterial cyclopropane synthase inhibitors. J Chem Sci 128, 719–732 (2016). https://doi.org/10.1007/s12039-016-1069-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12039-016-1069-1