Abstract
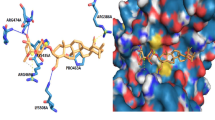
Cancer poses a significant global health challenge and significantly contributes to mortality. NEK7, related to the NIMA protein kinase family, plays a crucial role in spindle assembly and cell division. The dysregulation of NEK7 is closely linked to the onset and progression of various cancers, especially colon and breast cancer, making it a promising target for cancer therapy. Nevertheless, the shortage of high-quality NEK7 inhibitors highlights the need for new therapeutic strategies. In this study, we utilized a multidisciplinary approach, including virtual screening, molecular docking, pharmacokinetics, molecular dynamics simulations (MDs), and MM/PBSA calculations, to evaluate natural compounds as NEK7 inhibitors comprehensively. Through various docking strategies, we identified three natural compounds: (−)-balanol, digallic acid, and scutellarin. Molecular docking revealed significant interactions at residues such as GLU112 and ALA114, with docking scores of −15.054, −13.059, and −11.547 kcal/mol, respectively, highlighting their potential as NEK7 inhibitors. MDs confirmed the stability of these compounds at the NEK7-binding site. Hydrogen bond analysis during simulations revealed consistent interactions, supporting their strong binding capacity. MM/PBSA analysis identified other crucial amino acids contributing to binding affinity, including ILE20, VAL28, ILE75, LEU93, ALA94, LYS143, PHE148, LEU160, and THR161, crucial for stabilizing the complex. This research demonstrated that these compounds exceeded dabrafenib in binding energy, according to MM/PBSA calculations, underscoring their effectiveness as NEK7 inhibitors. ADME/T predictions showed lower oral toxicity for these compounds, suggesting their potential for further development. This study highlights the promise of these natural compounds as bases for creating more potent derivatives with significant biological activities, paving the way for future experimental validation.
















Similar content being viewed by others
Abbreviations
- ADME/T:
-
Absorption, distribution, metabolism, excretion, and toxicity
- CL:
-
Clearance
- DCCM:
-
Dynamic cross-correlation map
- DFT:
-
Density functional theory
- DSSP:
-
Definition secondary structure of protein
- FEL:
-
Free energy landscape
- FMO:
-
Frontier molecular orbitals
- H-bond:
-
Hydrogen bond
- HOMO:
-
Highest occupied molecular orbital
- LUMO:
-
Lowest unoccupied molecular orbital
- MDCK:
-
Madin–Darby canine kidney
- MDs:
-
Molecular dynamics simulations
- MEP:
-
Molecular electrostatic potential
- MM/PBSA:
-
Molecular mechanics Poisson–Boltzmann surface area
- NEK:
-
NIMA-related kinase
- PCA:
-
Principal component analysis
- PDB:
-
Protein database
- PKC:
-
Protein kinase C
- PPB:
-
Plasma protein binding
- Rg:
-
Radius of gyration
- RMSD:
-
Root-mean-square deviation
- RMSF:
-
Root-mean-square fluctuation
- SBVS:
-
Structure-based virtual screening
- SMILES:
-
Simplified Molecular Input Line Entry System
- T 1/2 :
-
Half-life
References
Nguyen K, Boehling J, Tran MN, Cheng T, Rivera A, Collins-Burow BM, Lee SB, Drewry DH, Burow ME (2023) NEK family review and correlations with patient survival outcomes in various cancer types. Cancers 15(7):2067
van de Kooij B, Creixell P, van Vlimmeren A, Joughin BA, Miller CJ, Haider N, Simpson CD, Linding R, Stambolic V, Turk BE, Yaffe MB (2019) Comprehensive substrate specificity profiling of the human Nek kinome reveals unexpected signaling outputs. Life 8:e44635. https://doi.org/10.7554/eLife.44635
Bachus S, Graves D, Fulham L, Akkerman N, Stephanson C, Shieh J, Pelka P (2022) In mitosis you are not: the NIMA family of kinases in Aspergillus, yeast, and mammals. Int J Mol Sci 23(7):4041
Yamamoto Y, Chino H, Tsukamoto S, Ode KL, Ueda HR, Mizushima N (2021) NEK9 regulates primary cilia formation by acting as a selective autophagy adaptor for MYH9/myosin IIA. Nat Commun 12(1):3292. https://doi.org/10.1038/s41467-021-23599-7
Panchal NK, Evan Prince S (2023) The NEK family of serine/threonine kinases as a biomarker for cancer. Clin Exp Med 23(1):17–30. https://doi.org/10.1007/s10238-021-00782-0
Au FKC, Hau BKT, Qi RZ (2020) Nek2-mediated GAS2L1 phosphorylation and centrosome-linker disassembly induce centrosome disjunction. J Cell Biol 219(5):e201909094. https://doi.org/10.1083/jcb.201909094
Gorry R, Brennan K, Lavin PTM, Sheridan R, McGee MM (2023) Phosphorylation of the prolyl isomerase Cyclophilin A regulates its localisation and release from the centrosome during mitosis. Cell Cycle 22(8):951–966. https://doi.org/10.1080/15384101.2023.2167430
Richards MW, O’Regan L, Mas-Droux C, Blot JMY, Cheung J, Hoelder S, Fry AM, Bayliss R (2009) An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol Cell 36(4):560–570. https://doi.org/10.1016/j.molcel.2009.09.038
Müller M, Eghbalian R, Boeckel J-N, Frese KS, Haas J, Kayvanpour E, Sedaghat-Hamedani F, Lackner MK, Tugrul OF, Ruppert T, Tappu R, Martins Bordalo D, Kneuer JM, Piekarek A, Herch S, Schudy S, Keller A, Grammes N, Bischof C, Klinke A, Cardoso-Moreira M, Kaessmann H, Katus HA, Frey N, Steinmetz LM, Meder B (2022) NIMA-related kinase 9 regulates the phosphorylation of the essential myosin light chain in the heart. Nat Commun 13(1):6209. https://doi.org/10.1038/s41467-022-33658-2
de Souza EE, Meirelles GV, Godoy BB, Perez AM, Smetana JHC, Doxsey SJ, McComb ME, Costello CE, Whelan SA, Kobarg J (2014) Characterization of the human NEK7 interactome suggests catalytic and regulatory properties distinct from those of NEK6. J Proteome Res 13(9):4074–4090. https://doi.org/10.1021/pr500437x
Freixo F, Martinez Delgado P, Manso Y, Sánchez-Huertas C, Lacasa C, Soriano E, Roig J, Lüders J (2018) NEK7 regulates dendrite morphogenesis in neurons via Eg5-dependent microtubule stabilization. Nat Commun 9(1):2330. https://doi.org/10.1038/s41467-018-04706-7
O’Regan L, Barone G, Adib R, Woo CG, Jeong HJ, Richardson EL, Richards MW, Muller PAJ, Collis SJ, Fennell DA, Choi J, Bayliss R, Fry AM (2020) EML4–ALK V3 oncogenic fusion proteins promote microtubule stabilization and accelerated migration through NEK9 and NEK7. J Cell Sci 133(9):jcs241505. https://doi.org/10.1242/jcs.241505
Eisa NH, Jilani Y, Kainth K, Redd P, Lu S, Bougrine O, Abdul Sater H, Patwardhan CA, Shull A, Shi H, Liu K, Elsherbiny NM, Eissa LA, El-Shishtawy MM, Horuzsko A, Bollag R, Maihle N, Roig J, Korkaya H, Cowell JK, Chadli A (2019) The co-chaperone UNC45A is essential for the expression of mitotic kinase NEK7 and tumorigenesis. J Biol Chem 294(14):5246–5260. https://doi.org/10.1074/jbc.RA118.006597
Kang D, Cho H-S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Hayami S, Tsunoda T, Field HI, Matsuda K, Neal DE, Ponder BAJ, Maehara Y, Nakamura Y, Hamamoto R (2013) The histone methyltransferase Wolf-Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosomes Cancer 52(2):126–139. https://doi.org/10.1002/gcc.22012
Saloura V, Cho H-S, Kiyotani K, Alachkar H, Zuo Z, Nakakido M, Tsunoda T, Seiwert T, Lingen M, Licht J, Nakamura Y, Hamamoto R (2015) WHSC1 promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol Cancer Res 13(2):293–304. https://doi.org/10.1158/1541-7786.Mcr-14-0292-t
Yan Z, Qu J, Li Z, Yi J, Su Y, Lin Q, Yu G, Lin Z, Yin W, Lu F, Liu J (2021) NEK7 promotes pancreatic cancer progression and its expression is correlated with poor prognosis. Front Oncol 11:705797. https://doi.org/10.3389/fonc.2021.705797
Sun Z, Gong W, Zhang Y, Jia Z (2020) Physiological and pathological roles of mammalian NEK7. Front Physiol 11:606996. https://doi.org/10.3389/fphys.2020.606996
Phadke M, Remsing Rix LL, Smalley I, Bryant AT, Luo Y, Lawrence HR, Schaible BJ, Chen YA, Rix U, Smalley KSM (2018) Dabrafenib inhibits the growth of BRAF-WT cancers through CDK16 and NEK9 inhibition. Mol Oncol 12(1):74–88. https://doi.org/10.1002/1878-0261.12152
Bagherniya M, Mahdavi A, Shokri-Mashhadi N, Banach M, Von Haehling S, Johnston TP, Sahebkar A (2022) The beneficial therapeutic effects of plant-derived natural products for the treatment of sarcopenia. J Cachexia Sarcopenia Muscle 13(6):2772–2790. https://doi.org/10.1002/jcsm.13057
Hernandez DF, Cervantes EL, Luna-Vital DA, Mojica L (2021) Food-derived bioactive compounds with anti-aging potential for nutricosmetic and cosmeceutical products. Crit Rev Food Sci Nutr 61(22):3740–3755. https://doi.org/10.1080/10408398.2020.1805407
Yu L, Tao J, Zhao Q, Xu C, Zhang Q (2020) Confirmation of potential neuroprotective effects of natural bioactive compounds from traditional medicinal herbs in cerebral ischemia treatment. J Integr Neurosci 19(2):373–384. https://doi.org/10.31083/j.jin.2020.02.63
Rampogu S, Lee G, Kulkarni AM, Kim D, Yoon S, Kim MO, Lee KW (2021) Computational approaches to discover novel natural compounds for SARS-CoV-2 therapeutics. ChemistryOpen 10(5):593–599. https://doi.org/10.1002/open.202000332
van Santen JA, Jacob G, Singh AL, Aniebok V, Balunas MJ, Bunsko D, Neto FC, Castaño-Espriu L, Chang C, Clark TN, Cleary Little JL, Delgadillo DA, Dorrestein PC, Duncan KR, Egan JM, Galey MM, Haeckl FPJ, Hua A, Hughes AH, Iskakova D, Khadilkar A, Lee J-H, Lee S, LeGrow N, Liu DY, Macho JM, McCaughey CS, Medema MH, Neupane RP, O’Donnell TJ, Paula JS, Sanchez LM, Shaikh AF, Soldatou S, Terlouw BR, Tran TA, Valentine M, van der Hooft JJJ, Vo DA, Wang M, Wilson D, Zink KE, Linington RG (2019) The natural products atlas: an open access knowledge base for microbial natural products discovery. ACS Cent Sci 5(11):1824–1833. https://doi.org/10.1021/acscentsci.9b00806
van Santen JA, Poynton EF, Iskakova D, McMann E, Alsup Tyler A, Clark TN, Fergusson CH, Fewer DP, Hughes AH, McCadden CA, Parra J, Soldatou S, Rudolf JD, Janssen EM-L, Duncan KR, Linington RG (2021) The Natural Products Atlas 2.0: a database of microbially-derived natural products. Nucleic Acids Res 50(D1):D1317–D1323. https://doi.org/10.1093/nar/gkab941
Irwin JJ, Tang KG, Young J, Dandarchuluun C, Wong BR, Khurelbaatar M, Moroz YS, Mayfield J, Sayle RA (2020) ZINC20—a free ultralarge-scale chemical database for ligand discovery. J Chem Inf Model 60(12):6065–6073. https://doi.org/10.1021/acs.jcim.0c00675
Sterling T, Irwin JJ (2015) ZINC 15—ligand discovery for everyone. J Chem Inf Model 55(11):2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Naz S, Ashraf S, Parvez MK, Al-Dosari MS, Ul-Haq Z (2022) Structure and ligand-based drug discovery of IL-4 inhibitors via interaction-energy-based learning approaches. J Biomol Struct Dyn 40(14):6503–6521. https://doi.org/10.1080/07391102.2021.1886172
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Xue Q, Liu X, Russell P, Li J, Pan W, Fu J, Zhang A (2022) Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock. Ecotoxicol Environ Saf 233:113323. https://doi.org/10.1016/j.ecoenv.2022.113323
Abdulwahab MK, Tan KH, Dzulkeflee R, Leong KH, Heh CH, Ariffin A (2021) In-silico studies of the antiproliferative activity of new anilinoquinazoline derivatives against NSCLC cells. J Mol Struct 1228:129786. https://doi.org/10.1016/j.molstruc.2020.129786
Perike N, Edigi PK, Nirmala G, Thumma V, Bujji S, Naikal PS (2022) Synthesis, anticancer activity and molecular docking studies of hybrid molecules containing indole-thiazolidinedione-triazole moieties. ChemistrySelect 7(47):e202203778. https://doi.org/10.1002/slct.202203778
Bell EW, Zhang Y (2019) DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J Cheminformatics 11(1):40. https://doi.org/10.1186/s13321-019-0362-7
Stanzione F, Giangreco I, Cole JC (2021) Chapter 4—Use of molecular docking computational tools in drug discovery. In: Witty DR, Cox B (eds) Progress in medicinal chemistry. Elsevier, pp 273–343
Jablonský M, Štekláč M, Majová V, Gall M, Matúška J, Pitoňák M, Bučinský L (2022) Molecular docking and machine learning affinity prediction of compounds identified upon softwood bark extraction to the main protease of the SARS-CoV-2 virus. Biophys Chem 288:106854. https://doi.org/10.1016/j.bpc.2022.106854
Aziz M, Ejaz SA, Rehman HM, Alsubaie AS, Mahmoud KH, Siddique F, Al-Buriahi MS, Alrowaili ZA (2022) Identification of NEK7 inhibitors: structure based virtual screening, molecular docking, density functional theory calculations and molecular dynamics simulations. J Biomol Struct Dyn 41(14):6894–6908. https://doi.org/10.1080/07391102.2022.2113563
Safna Hussan KP, Abdul Rahoof KA, Medammal Z, Thayyil MS, Babu TD (2022) Theoretical insights into the radical scavenging activity of glipizide: DFT and molecular docking studies. Free Radic Res 56(1):53–62. https://doi.org/10.1080/10715762.2022.2034803
Siddiqui SA (2020) In silico design of organic p–n junction diodes using quantum chemical calculations. J Comput Electron 19(1):80–90. https://doi.org/10.1007/s10825-020-01447-z
Arivazhagan R, Sridevi C, Prakasam A (2021) Exploring molecular structure, spectral features, electronic properties and molecular docking of a novel biologically active heterocyclic compound 4-phenylthiosemicarbazide. J Mol Struct 1232:129956. https://doi.org/10.1016/j.molstruc.2021.129956
Sinha P, Yadav AK (2023) Identification of 3,4-dihydroxy complexes as potential antiviral via DFT, molecular docking, molecular dynamics and MM/PBSA against rabies and dengue receptors. J Biomol Struct Dyn 2023:1–17. https://doi.org/10.1080/07391102.2023.2246572
Lin Y, Zhang Y, Wang D, Yang B, Shen Y-Q (2022) Computer especially AI-assisted drug virtual screening and design in traditional Chinese medicine. Phytomedicine 107:154481. https://doi.org/10.1016/j.phymed.2022.154481
Belghalia E, Ouabane M, El Bahi S, Rehman HM, Sbai A, Lakhlifi T, Bouachrine M (2023) In silico research on new sulfonamide derivatives as BRD4 inhibitors targeting acute myeloid leukemia using various computational techniques including 3D-QSAR, HQSAR, molecular docking, ADME/Tox, and molecular dynamics. J Biomol Struct Dyn 2023:1–19. https://doi.org/10.1080/07391102.2023.2250460
Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49(W1):W5–W14. https://doi.org/10.1093/nar/gkab255
Zhang D, Wang Z, Li J, Zhu J (2022) Exploring the possible molecular targeting mechanism of Saussurea involucrata in the treatment of COVID-19 based on bioinformatics and network pharmacology. Comput Biol Med 146:105549. https://doi.org/10.1016/j.compbiomed.2022.105549
Che Omar MT (2020) Data analysis of molecular dynamics simulation trajectories of β-sitosterol, sonidegib and cholesterol in smoothened protein with the CHARMM36 force field. Data Brief 33:106350. https://doi.org/10.1016/j.dib.2020.106350
Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26(16):1668–1688. https://doi.org/10.1002/jcc.20290
Sapay N, Tieleman DP (2011) Combination of the CHARMM27 force field with united-atom lipid force fields. J Comput Chem 32(7):1400–1410. https://doi.org/10.1002/jcc.21726
Stroet M, Caron B, Visscher KM, Geerke DP, Malde AK, Mark AE (2018) Automated Topology Builder Version 3.0: prediction of solvation free enthalpies in water and hexane. J Chem Theory Comput 14(11):5834–5845. https://doi.org/10.1021/acs.jctc.8b00768
Moharana M, Pattanayak SK, Khan F (2022) Computational efforts to identify natural occurring compounds from Phyllanthus niruri that target hepatitis B viral infections: DFT, docking and dynamics simulation study. J Indian Chem Soc 99(9):100662. https://doi.org/10.1016/j.jics.2022.100662
Abdi H, Williams LJ (2010) Principal component analysis. WIREs Comput Statistics 2(4):433–459. https://doi.org/10.1002/wics.101
Ringnér M (2008) What is principal component analysis? Nat Biotechnol 26(3):303–304. https://doi.org/10.1038/nbt0308-303
Srikumar PS, Rohini K, Rajesh PK (2014) Molecular dynamics simulations and principal component analysis on human laforin mutation W32G and W32G/K87A. Protein J 33(3):289–295. https://doi.org/10.1007/s10930-014-9561-2
Swain SS, Paidesetty SK, Dehury B, Sahoo J, Vedithi SC, Mahapatra N, Hussain T, Padhy RN (2018) Molecular docking and simulation study for synthesis of alternative dapsone derivative as a newer antileprosy drug in multidrug therapy. J Cell Biochem 119(12):9838–9852. https://doi.org/10.1002/jcb.27304
Ji H-F, Shen L, Grandori R, Müller N (2008) The effect of heme on the conformational stability of micro-myoglobin. FEBS J 275(1):89–96. https://doi.org/10.1111/j.1742-4658.2007.06176.x
Ramachandrakurup S, Ramakrishnan V (2017) Effect of NaeI-L43K mutation on protein dynamics and DNA conformation: insights from molecular dynamics simulations. J Mol Graph Model 76:456–465. https://doi.org/10.1016/j.jmgm.2017.07.029
Torabi R, Bagherzadeh K, Ghourchian H, Amanlou M (2016) An investigation on the interaction modes of a single-strand DNA aptamer and RBP4 protein: a molecular dynamic simulations approach. Electronic supplementary information (ESI) available. Org Biomol Chem 14(34):8141–8153. https://doi.org/10.1039/c6ob01094f
Cembran A, Masterson LR, McClendon CL, Taylor SS, Gao J, Veglia G (2012) Conformational equilibrium of N-myristoylated cAMP-dependent protein kinase A by molecular dynamics simulations. Biochemistry 51(51):10186–10196. https://doi.org/10.1021/bi301279f
Chillemi G, D’Annessa I, Fiorani P, Losasso C, Benedetti P, Desideri A (2008) Thr729 in human topoisomerase I modulates anti-cancer drug resistance by altering protein domain communications as suggested by molecular dynamics simulations. Nucleic Acids Res 36(17):5645–5651. https://doi.org/10.1093/nar/gkn558
Zacharias J, Knapp E-W (2014) Protein secondary structure classification revisited: processing DSSP information with PSSC. J Chem Inf Model 54(7):2166–2179. https://doi.org/10.1021/ci5000856
Zhang Y, Sagui C (2015) Secondary structure assignment for conformationally irregular peptides: comparison between DSSP, STRIDE and KAKSI. J Mol Graph Model 55:72–84. https://doi.org/10.1016/j.jmgm.2014.10.005
King E, Aitchison E, Li H, Luo R (2021) Recent developments in free energy calculations for drug discovery. Front Mol Biosci 8:712085. https://doi.org/10.3389/fmolb.2021.712085
Poli G, Granchi C, Rizzolio F, Tuccinardi T (2020) Application of MM-PBSA methods in virtual screening. Molecules 25(8):1971
Samanta R, Pradhan KK, Sen D, Kar S, Ghosh M (2023) Structure-based drug design-guided identification of estrogen receptor binders. Mol Divers. https://doi.org/10.1007/s11030-023-10657-z
Hevener KE, Zhao W, Ball DM, Babaoglu K, Qi J, White SW, Lee RE (2009) Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model 49(2):444–460. https://doi.org/10.1021/ci800293n
Vesaghhamedani S, Mazloumi Kiapey SS, Gowhari Shabgah A, Amiresmaili S, Jahanara A, Oveisee M, Shekarchi A, Gheibihayat SM, Jadidi-Niaragh F, Gholizadeh Navashenaq J (2023) From traditional medicine to modern oncology: scutellarin, a promising natural compound in cancer treatment. Prog Biophys Mol Biol 180–181:19–27. https://doi.org/10.1016/j.pbiomolbio.2023.04.006
Alfonso EE, Troche R, Deng Z, Annamalai T, Chapagain P, Tse-Dinh Y-C, Leng F (2022) Potent inhibition of bacterial DNA gyrase by digallic acid and other gallate derivatives. ChemMedChem 17(23):e202200301. https://doi.org/10.1002/cmdc.202200301
Bhouri W, Boubaker J, Skandrani I, Ghedira K, Chekir Ghedira L (2012) Investigation of the apoptotic way induced by digallic acid in human lymphoblastoid TK6 cells. Cancer Cell Int 12(1):26. https://doi.org/10.1186/1475-2867-12-26
Hardianto A, Yusuf M, Liu F, Ranganathan S (2017) Exploration of charge states of balanol analogues acting as ATP-competitive inhibitors in kinases. BMC Bioinformatics 18(16):572. https://doi.org/10.1186/s12859-017-1955-7
Nogara PA, Saraiva RdA, Caeran Bueno D, Lissner LJ, Lenz Dalla Corte C, Braga MM, Rosemberg DB, Rocha JBT (2015) Virtual screening of acetylcholinesterase inhibitors using the Lipinski’s rule of five and ZINC Databank. Biomed Res Int 2015:870389. https://doi.org/10.1155/2015/870389
O’Donovan DH, De Fusco C, Kuhnke L, Reichel A (2023) Trends in molecular properties, bioavailability, and permeability across the Bayer compound collection. J Med Chem 66(4):2347–2360. https://doi.org/10.1021/acs.jmedchem.2c01577
Hu L, Wang J, Zhao X, Cai D (2022) Mechanism of saikogenin G against major depressive disorder determined by network pharmacology. Medicine 101(34):e30193. https://doi.org/10.1097/md.0000000000030193
El Aissouq A, Bouachrine M, Ouammou A, Khalil F (2022) Homology modeling, virtual screening, molecular docking, molecular dynamic (MD) simulation, and ADMET approaches for identification of natural anti-Parkinson agents targeting MAO-B protein. Neurosci Lett 786:136803. https://doi.org/10.1016/j.neulet.2022.136803
Roy R, Sk MF, Jonniya NA, Poddar S, Kar P (2022) Finding potent inhibitors against SARS-CoV-2 main protease through virtual screening, ADMET, and molecular dynamics simulation studies. J Biomol Struct Dyn 40(14):6556–6568. https://doi.org/10.1080/07391102.2021.1897680
Almazroo OA, Miah MK, Venkataramanan R (2017) Drug metabolism in the liver. Clin Liver Dis 21(1):1–20. https://doi.org/10.1016/j.cld.2016.08.001
Zhou S, Zhao F-L, Wang S-H, Wang Y-R, Hong Y, Zhou Q, Geng P-W, Luo Q-F, Cai J-P, Dai D-P (2023) Assessments of CYP-inhibition-based drug–drug interaction between vonoprazan and poziotinib in vitro and in vivo. Pharm Biol 61(1):356–361. https://doi.org/10.1080/13880209.2023.2173253
Islam MR, Osman OI, Hassan WMI (2023) Identifying novel therapeutic inhibitors to target FMS-like tyrosine kinase-3 (FLT3) against acute myeloid leukemia: a molecular docking, molecular dynamics, and DFT study. J Biomol Struct Dyn 2023:1–19. https://doi.org/10.1080/07391102.2023.2192798
Su A, Zhang X, Zhang C, Ding D, Yang Y-F, Wang K, She Y-B (2023) Deep transfer learning for predicting frontier orbital energies of organic materials using small data and its application to porphyrin photocatalysts. Phys Chem Chem Phys 25(15):10536–10549. https://doi.org/10.1039/D3CP00917C
Arshad R, Khan MA, Mutahir S, Hussain S, Al-Hazmi GH, Refat MS (2022) DFT, molecular docking and ADME studies of thiazolidinones as tyrosinase inhibitors. Polycyclic Arom Compounds 43:6750–6765. https://doi.org/10.1080/10406638.2022.2124286
Ashok AK, Gnanasekaran TS, Santosh Kumar HS, Srikanth K, Prakash N, Gollapalli P (2023) High-throughput screening and molecular dynamics simulations of natural products targeting LuxS/AI-2 system as a novel antibacterial strategy for antibiotic resistance in Helicobacter pylori. J Biomol Struct Dyn 42(6):2913–2928. https://doi.org/10.1080/07391102.2023.2210674
Ashiru MA, Ogunyemi SO, Temionu OR, Ajibare AC, Cicero-Mfon NC, Ihekuna OA, Jagun MO, Abdulmumin L, Adisa QK, Asibor YE, Okorie CJ, Lawal MO, Babalola MO, Abdulrasaq IT, Salau LB, Olatunji IO, Bankole MA, Daud AB, Adeyemi AO (2023) Identification of EGFR inhibitors as potential agents for cancer therapy: pharmacophore-based modeling, molecular docking, and molecular dynamics investigations. J Mol Model 29(5):128. https://doi.org/10.1007/s00894-023-05531-6
Guterres H, Park S-J, Jiang W, Im W (2021) Ligand-binding-site refinement to generate reliable holo protein structure conformations from apo structures. J Chem Inf Model 61(1):535–546. https://doi.org/10.1021/acs.jcim.0c01354
Singh A, Mishra A (2022) Molecular modelling study to discover novel JAK2 signaling pathway inhibitor. J Biomol Struct Dyn 41(12):5827–5838. https://doi.org/10.1080/07391102.2022.2097314
Chen J, Yu X, Chen Q, Wu Q, He Q (2022) Screening and mechanisms of novel angiotensin-I-converting enzyme inhibitory peptides from rabbit meat proteins: a combined in silico and in vitro study. Food Chem 370:131070. https://doi.org/10.1016/j.foodchem.2021.131070
Hidayatullah A, Putra WE, Widiastuti D, Salma WO, Heikal MF (2023) Molecular docking and molecular dynamics simulation-based identification of natural inhibitors against druggable human papilloma virus type 16 target. Trends Sci 20(4):4891. https://doi.org/10.48048/tis.2023.4891
Almeleebia TM, Ahamad S, Ahmad I, Alshehri A, Alkhathami AG, Alshahrani MY, Asiri MA, Saeed A, Siddiqui JA, Yadav DK, Saeed M (2022) Identification of PARP12 inhibitors by virtual screening and molecular dynamics simulations. Front Pharmacol 13:847499. https://doi.org/10.3389/fphar.2022.847499
Kumar DT, Shaikh N, Kumar SU, Doss CGP, Zayed H (2021) Structure-based virtual screening to identify novel potential compound as an alternative to remdesivir to overcome the RdRp protein mutations in SARS-CoV-2. Front Mol Biosci 8:645216. https://doi.org/10.3389/fmolb.2021.645216
Shafie A, Khan S, Zehra MT, Anjum F, Hasan GM, Yadav DK, Hassan MI (2021) Identification of phytoconstituents as potent inhibitors of casein kinase-1 alpha using virtual screening and molecular dynamics simulations. Pharmaceutics 13(12):2157
Markthaler D, Fleck M, Stankiewicz B, Hansen N (2022) Exploring the effect of enhanced sampling on protein stability prediction. J Chem Theory Comput 18(4):2569–2583. https://doi.org/10.1021/acs.jctc.1c01012
Borkotoky S, Banerjee M, Modi GP, Dubey VK (2021) Identification of high affinity and low molecular alternatives of boceprevir against SARS-CoV-2 main protease: a virtual screening approach. Chem Phys Lett 770:138446. https://doi.org/10.1016/j.cplett.2021.138446
Funding
The author(s) reported there is no funding associated with the work featured in this article.
Author information
Authors and Affiliations
Contributions
HZ and CL conceptualized the study and designed the research methodology. HZ and QY performed the virtual screening, molecular docking, and ADME/T predictions. CL and QJ handled molecular dynamics (MD) simulations, molecular mechanics Poisson-Boltzmann surface area (MM/PBSA)-based binding free energy calculations, and DFT calculations. HZ and CL analyzed the results and wrote the initial draft of the manuscript. QJ prepared the figures and tables. QY provided critical revisions to the manuscript for important intellectual content. All authors participated in the review, editing process, and approved the final manuscript for publication.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, H., Lu, C., Yao, Q. et al. In silico study to identify novel NEK7 inhibitors from natural sources by a combination strategy. Mol Divers (2024). https://doi.org/10.1007/s11030-024-10838-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-024-10838-4