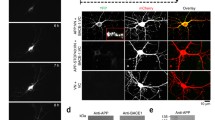
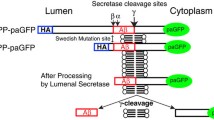
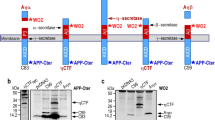
Abstract
Background
Alzheimer’s disease amyloid-beta peptides (Aβ) are generated via sequential cleavage of the amyloid precursor protein (APP) by β-secretase (Bace1) and γ-secretase. Though the precise subcellular location(s) of Bace1-mediated APP cleavage remains unresolved, current models suggest APP internalization into Bace1-containing endosomes is a critical step. However, direct evidence for this model is lacking, and previous reports that probed the APP/Bace1 interaction (using co-expressed APP and Bace1 differentially labeled with fluorescent protein tags) did not determine if APP fluorescence originated from full-length APP (fl-APP) molecules that had internalized from the cell surface pool.
Methods
We adapted the bungarotoxin-ligand (BTX) system to label surface APP and track internalized fluorescent APP/BTX puncta in rodent primary neurons co-expressing fluorescently-tagged Bace1. Subsequently, we employed imaging and biochemical-based approaches to measure N- and C-terminal APP epitope levels in primary neurons, N2a neuroblastoma, and HeLa cell lines.
Results
We hypothesized that surface-labeled APP/BTX puncta would, upon internalization, colocalize with fluorescently-tagged Bace1. Unexpectedly, we observed a dramatic loss of internalized APP in co-transfected neurons and ~ 80–90% loss of surface-resident fl-APP, which we also observed in HeLa and N2a cells. Loss of surface fl-APP could be reversed by a Bace1 inhibitor, suggesting that enhanced Bace1-mediated APP cleavage was responsible for the altered processing and mis-sorting. Importantly, in a C-terminally-tagged APP construct, the majority of C-terminal fluorescence was preserved in HeLa cells despite the loss of N-terminal APP signal. This phenomenon was not only recapitulated in cultured neurons, but also showed a progressive disappearance of the APP N-terminal tag, reflecting continual cleavage of fl-APP by Bace1 away from the cell body.
Conclusions
Our results strongly suggested that in APP/Bace1 co-expression approaches, there was significant early and aberrant Bace1-mediated APP cleavage that perturbed fl-APP trafficking from the secretory pathway onwards, resulting in a substantial loss of surface fl-APP, which in turn led to a marked reduction in APP internalization. In C-terminally-tagged APP constructs, a large fraction of the APP fluorescence signal therefore likely arose from fluorescently-tagged β-C-terminal-fragment (β-CTF) or downstream proteolytic derivatives instead of fl-APP. Thus, care is needed in interpreting results where APP is detected only with a C-terminal tag in the presence of Bace1 co-expression, and previous findings may need to be reinterpreted if it is unclear whether fl-APP is present in normal physiological levels.







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in the published article [and its supplementary information files].
Abbreviations
- β-CTF:
-
β-C-terminal fragment
- A647:
-
Alexa-fluor-647
- Aβ:
-
Amyloid-beta
- AD:
-
Alzheimer’s disease
- AICD:
-
APP intracellular domain
- APP:
-
Amyloid precursor protein
- APP-CTF:
-
APP-C-terminal-fragment
- Βace1:
-
β-Secretase
- BBS:
-
α-Bungarotoxin binding site
- BFP:
-
Blue fluorescent protein
- BSI:
-
Bace1-inhibitor
- BTX:
-
α-Bungarotoxin
- CME:
-
Clathrin-/dynamin-mediated endocytosis
- DIV:
-
Days-in-vitro
- FBS:
-
Fetal bovine serum
- FL-APP:
-
Full-length APP
- FRET:
-
Forster resonance energy transfers
- FWHM:
-
Full-width half-maximum
- GFP:
-
Green fluorescent protein
- HRP:
-
Horse-radish peroxidase
- mCh:
-
MCherry
- MEM-NEAA:
-
Minimum Essential Medium non-essential amino acids
- Nb-A:
-
Neurobasal-A
- NH4Cl:
-
Ammonium chloride
- PBS:
-
Phosphate-buffered saline
- PDL:
-
Poly-d-lysine
- PFA:
-
Paraformaldehyde
- RFP:
-
Red fluorescent protein
- ROIs:
-
Regions-of-interest
- sAPPβ:
-
Soluble-APP-β fragment
- SEM:
-
Standard error of measurement
- SEP:
-
Super-ecliptic pHluorin
- TBST:
-
Tris-buffered saline with Tween
- TGN:
-
Trans-Golgi-network
References
Selkoe DJ, Hardy J (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8(6):595–608
Nhan HS, Chiang K, Koo EH (2015) The multifaceted nature of amyloid precursor protein and its proteolytic fragments: friends and foes. Acta Neuropathol 129(1):1–19
Haass C, Kaether C, Thinakaran G, Sisodia S (2012) Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med. 2(5):a006270
Rajendran L, Annaert W (2012) Membrane trafficking pathways in Alzheimer’s disease. Traffic 13(6):759–770
Toh WH, Gleeson PA (2016) Dysregulation of intracellular trafficking and endosomal sorting in Alzheimer’s disease: controversies and unanswered questions. Biochem J 473(14):1977–1993
Toh WH, Tan JZ, Zulkefli KL, Houghton FJ, Gleeson PA (2017) Amyloid precursor protein traffics from the Golgi directly to early endosomes in an Arl5b- and AP4-dependent pathway. Traffic 18(3):159–175
Tam JH, Seah C, Pasternak SH (2014) The Amyloid Precursor Protein is rapidly transported from the Golgi apparatus to the lysosome and where it is processed into beta-amyloid. Mol Brain 7:54
Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW et al (2010) ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J 29(17):3020–3032
Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P et al (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286(5440):735–741
Cole SL, Vassar R (2007) The Basic Biology of BACE1: A Key Therapeutic Target for Alzheimer’s Disease. Curr Genomics 8(8):509–530
Sathya M, Premkumar P, Karthick C, Moorthi P, Jayachandran KS, Anusuyadevi M (2012) BACE1 in Alzheimer’s disease. Clin Chim Acta 414:171–178
Yan R, Vassar R (2014) Targeting the β-secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol 13(3):319–329
Tan J, Evin G (2012) β-site APP-cleaving enzyme 1 trafficking and Alzheimer’s disease pathogenesis. J Neurochem 120(6):869–880
Hemming ML, Elias JE, Gygi SP, Selkoe DJ (2009) Identification of beta-secretase (BACE1) substrates using quantitative proteomics. PLoS One. 4(12):e8477
Ben Halima S, Mishra S, Raja KMP, Willem M, Baici A, Simons K et al (2016) Specific Inhibition of β-Secretase Processing of the Alzheimer Disease Amyloid Precursor Protein. Cell Rep 14(9):2127–2141
Zhang X, Song W (2013) The role of APP and BACE1 trafficking in APP processing and amyloid-β generation. Alzheimers Res Ther 5(5):46
Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ et al (1999) Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem 274(27):18851–18856
Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA et al (2007) The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci 27(29):7817–7826
Carey RM, Balcz BA, Lopez-Coviella I, Slack BE (2005) Inhibition of dynamin-dependent endocytosis increases shedding of the amyloid precursor protein ectodomain and reduces generation of amyloid beta protein. BMC Cell Biol 6:30
Zhu L, Su M, Lucast L, Liu L, Netzer WJ, Gandy SE et al (2012) Dynamin 1 regulates amyloid generation through modulation of BACE-1. PLoS One. 7(9):e45033
Cirrito JR, Kang JE, Lee J, Stewart FR, Verges DK, Silverio LM et al (2008) Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron 58(1):42–51
Das U, Scott DA, Ganguly A, Koo EH, Tang Y, Roy S (2013) Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron 79(3):447–460
von Arnim CA, von Einem B, Weber P, Wagner M, Schwanzar D, Spoelgen R et al (2008) Impact of cholesterol level upon APP and BACE proximity and APP cleavage. Biochem Biophys Res Commun 370(2):207–212
Marquer C, Devauges V, Cossec JC, Liot G, Lécart S, Saudou F et al (2011) Local cholesterol increase triggers amyloid precursor protein-Bace1 clustering in lipid rafts and rapid endocytosis. FASEB J 25(4):1295–1305
Das U, Wang L, Ganguly A, Saikia JM, Wagner SL, Koo EH et al (2016) Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat Neurosci 19(1):55–64
Kinoshita A, Fukumoto H, Shah T, Whelan CM, Irizarry MC, Hyman BT (2003) Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci 116(Pt 16):3339–3346
von Einem B, Schwanzar D, Rehn F, Beyer AS, Weber P, Wagner M et al (2010) The role of low-density receptor-related protein 1 (LRP1) as a competitive substrate of the amyloid precursor protein (APP) for BACE1. Exp Neurol 225(1):85–93
von Einem B, Weber P, Wagner M, Malnar M, Kosicek M, Hecimovic S et al (2012) Cholesterol-dependent energy transfer between fluorescent proteins-insights into protein proximity of APP and BACE1 in different membranes in Niemann-Pick type C disease cells. Int J Mol Sci 13(12):15801–15812
Sekine-Aizawa Y, Huganir RL (2004) Imaging of receptor trafficking by using alpha-bungarotoxin-binding-site-tagged receptors. Proc Natl Acad Sci U S A 101(49):17114–17119
Mammen AL, Huganir RL, O’Brien RJ (1997) Redistribution and stabilization of cell surface glutamate receptors during synapse formation. J Neurosci 17(19):7351–7358
Hu YB, Dammer EB, Ren RJ, Wang G (2015) The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener 4:18
Kopec CD, Li B, Wei W, Boehm J, Malinow R (2006) Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J Neurosci 26(7):2000–2009
Muresan V, Varvel NH, Lamb BT, Muresan Z (2009) The cleavage products of amyloid-beta precursor protein are sorted to distinct carrier vesicles that are independently transported within neurites. J Neurosci 29(11):3565–3578
Sisodia SS (1992) Beta-amyloid precursor protein cleavage by a membrane-bound protease. Proc Natl Acad Sci U S A 89(13):6075–6079
Parvathy S, Hussain I, Karran EH, Turner AJ, Hooper NM (1999) Cleavage of Alzheimer’s amyloid precursor protein by alpha-secretase occurs at the surface of neuronal cells. Biochemistry 38(30):9728–9734
Small SA, Petsko GA (2015) Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat Rev Neurosci 16(3):126–132
Lee EB, Zhang B, Liu K, Greenbaum EA, Doms RW, Trojanowski JQ et al (2005) BACE overexpression alters the subcellular processing of APP and inhibits Abeta deposition in vivo. J Cell Biol 168(2):291–302
Caudano F, Montalto G, Passalacqua M, Pronzato MA, Fedele E, Ricciarelli R (2020) cGMP favors the interaction between APP and BACE1 by inhibiting Rab5 GTPase activity. Sci Rep 10(1):1358
Corish P, Tyler-Smith C (1999) Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 12(12):1035–1040
Bustamante HA, Rivera-Dictter A, Cavieres VA, Muñoz VC, González A, Lin Y et al (2013) Turnover of C99 is controlled by a crosstalk between ERAD and ubiquitin-independent lysosomal degradation in human neuroglioma cells. PLoS One. 8(12):e83096
Burgos PV, Mardones GA, Rojas AL, daSilva LL, Prabhu Y, Hurley JH et al (2010) Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev Cell 18(3):425–436
Kaether C, Schmitt S, Willem M, Haass C (2006) Amyloid precursor protein and Notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic 7(4):408–415
Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T et al (2003) APP processing and synaptic function. Neuron 37(6):925–937
Greicius MD, Srivastava G, Reiss AL, Menon V (2004) Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A 101(13):4637–4642
Acknowledgements
We thank Dr. Utpal Das and Dr. Subhojit Roy for providing the APP-GFP and Bace1-mCherry plasmids. We gratefully acknowledge members of the Koo lab for technical and administrative assistance, as well as for many helpful discussions and advice. We would also like to acknowledge the National University of Singapore Centre for Bioimaging Sciences (NUS-CBIS) for use of their confocal spinning disk microscopy system.
Funding
This work was funded in part by grants to EHK from the Singapore National Medical Research Council (NMRC, NMRC/StaR/009/2012) and the National Institutes of Health (NIH, NS84324). GT is funded by grants AG054223 and AG056061 from the National Institutes of Health. JA is funded by an Open-Fund Young Investigator’s Research Grant from NMRC (OF-YIRG, MOH-000237–00) and the Genome Institute of Singapore. TRH is supported by an Integrated Science and Engineering Programme scholarship from the National University of Singapore.
Author information
Authors and Affiliations
Contributions
JA, TRH and EK devised the project and planned experiments. JA and TRH carried out experiments and analyzed data. JA and TRH wrote the first draft of the manuscript. EK supervised the project and edited the manuscript with input from GT. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experiments were performed with the National University of Singapore Institutional Animal Care and Use Committee (IACUC) approval.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aow, J., Huang, TR., Thinakaran, G. et al. Enhanced cleavage of APP by co-expressed Bace1 alters the distribution of APP and its fragments in neuronal and non-neuronal cells. Mol Neurobiol 59, 3073–3090 (2022). https://doi.org/10.1007/s12035-022-02733-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02733-6