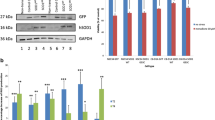
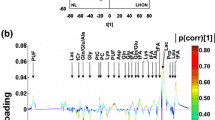
Abstract
Amyotrophic lateral sclerosis (ALS) is characterized by a wide metabolic remodeling, as shown by recent metabolomics and lipidomics studies performed in samples from patient cohorts and experimental animal models. Here, we explored the metabolome and lipidome of fibroblasts from sporadic ALS patients (n = 13) comparatively to age- and sex-matched controls (n = 11), and the subcellular fraction containing the mitochondria and endoplasmic reticulum (mito-ER), given that mitochondrial dysfunctions and ER stress are important features of ALS patho-mechanisms. We also assessed the mitochondrial oxidative respiration and the mitochondrial genomic (mtDNA) sequence, although without yielding significant differences. Compared to controls, ALS fibroblasts did not exhibit a mitochondrial respiration defect nor an increased proportion of mitochondrial DNA mutations. In addition, non-targeted metabolomics and lipidomics analyses identified 124 and 127 metabolites, and 328 and 220 lipids in whole cells and the mito-ER fractions, respectively, along with partial least-squares–discriminant analysis (PLS-DA) models being systematically highly predictive of the disease. The most discriminant metabolomic features were the alteration of purine, pyrimidine, and energetic metabolisms, suggestive of oxidative stress and of pro-inflammatory status. The most important lipidomic feature in the mito-ER fraction was the disturbance of phosphatidylcholine PC (36:4p) levels, which we had previously reported in the cerebrospinal fluid of ALS patients and in the brain from an ALS mouse model. Thus, our results reveal that fibroblasts from sporadic ALS patients share common metabolic remodeling, consistent with other metabolic studies performed in ALS, opening perspectives for further exploration in this cellular model in ALS.




Similar content being viewed by others
Abbreviations
- ALS:
-
amyotrophic lateral sclerosis
- sALS:
-
sporadic ALS
- ER:
-
endoplasmic reticulum
- mtDNA:
-
mitochondrial DNA
- PCA:
-
principal component analysis
- PLS-DA:
-
partial least square discriminant analysis
- OXPHOS:
-
oxidative phosphorylation
- FVC:
-
forced vital capacity
- BMI:
-
body mass index
- HRMS:
-
high-resolution mass spectrometry
- UPLC:
-
ultra-performance liquid chromatography
- ESI:
-
electrospray ionization
- QC:
-
quality control
- VIP:
-
variable importance in projection
- OCR:
-
oxygen consumption rate
- RCR:
-
respiration capacity rate
- SM:
-
sphingomyelins
- PC:
-
phosphatidylcholines
- PE:
-
phosphatidylethanolamines
- ROS:
-
reactive oxygen species
- CSF:
-
cerebrospinal fluid
- Cer:
-
ceramides
- DHA:
-
docosahexaenoic acid
References
Korner S, Kollewe K, Ilsemann J, Muller-Heine A, Dengler R, Krampfl K, Petri S (2013) Prevalence and prognostic impact of comorbidities in amyotrophic lateral sclerosis. Eur J Neurol 20(4):647–654. https://doi.org/10.1111/ene.12015
Barber SC, Shaw PJ (2010) Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic Biol Med 48(5):629–641. https://doi.org/10.1016/j.freeradbiomed.2009.11.018
Guegan C, Vila M, Rosoklija G, Hays AP, Przedborski S (2001) Recruitment of the mitochondrial-dependent apoptotic pathway in amyotrophic lateral sclerosis. J Neurosci 21(17):6569–6576
Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD (2001) Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis 8(6):933–941. https://doi.org/10.1006/nbdi.2001.0443
Ferraiuolo L, Kirby J, Grierson AJ, Sendtner M, Shaw PJ (2011) Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol 7(11):616–630. https://doi.org/10.1038/nrneurol.2011.152
Vandoorne T, De Bock K, Van Den Bosch L (2018) Energy metabolism in ALS: an underappreciated opportunity? Acta Neuropathol 135(4):489–509. https://doi.org/10.1007/s00401-018-1835-x
Konrad C, Kawamata H, Bredvik KG, Arreguin AJ, Cajamarca SA, Hupf JC, Ravits JM, Miller TM et al (2017) Fibroblast bioenergetics to classify amyotrophic lateral sclerosis patients. Mol Neurodegener 12(1):76. https://doi.org/10.1186/s13024-017-0217-5
Walczak J, Debska-Vielhaber G, Vielhaber S, Szymanski J, Charzynska A, Duszynski J, Szczepanowska J (2018) Distinction of sporadic and familial forms of ALS based on mitochondrial characteristics. Faseb j:fj201801843R. doi:https://doi.org/10.1096/fj.201801843R
Allen SP, Duffy LM, Shaw PJ, Grierson AJ (2015) Altered age-related changes in bioenergetic properties and mitochondrial morphology in fibroblasts from sporadic amyotrophic lateral sclerosis patients. Neurobiol Aging 36(10):2893–2903. https://doi.org/10.1016/j.neurobiolaging.2015.07.013
Allen SP, Rajan S, Duffy L, Mortiboys H, Higginbottom A, Grierson AJ, Shaw PJ (2014) Superoxide dismutase 1 mutation in a cellular model of amyotrophic lateral sclerosis shifts energy generation from oxidative phosphorylation to glycolysis. Neurobiol Aging 35(6):1499–1509. https://doi.org/10.1016/j.neurobiolaging.2013.11.025
Kirk K, Gennings C, Hupf JC, Tadesse S, D’Aurelio M, Kawamata H, Valsecchi F, Mitsumoto H et al (2014) Bioenergetic markers in skin fibroblasts of sporadic amyotrophic lateral sclerosis and progressive lateral sclerosis patients. Ann Neurol 76(4):620–624. https://doi.org/10.1002/ana.24244
Fonteh AN, Fisher RD (2009) Combining lipidomics and proteomics of human cerebrospinal fluids. Methods Mol Biol 579:71–86. https://doi.org/10.1007/978-1-60761-322-0_4
Puentes F, Malaspina A, van Noort JM, Amor S (2016) Non-neuronal cells in ALS: Role of glial, immune cells and blood-CNS barriers. Brain Pathol 26(2):248–257. https://doi.org/10.1111/bpa.12352
Szymanski J, Janikiewicz J, Michalska B, Patalas-Krawczyk P, Perrone M, Ziolkowski W, Duszynski J, Pinton P et al (2017) Interaction of mitochondria with the endoplasmic reticulum and plasma membrane in calcium homeostasis, lipid trafficking and mitochondrial structure. Int J Mol Sci 18(7). https://doi.org/10.3390/ijms18071576
Kaus A, Sareen D (2015) ALS patient stem cells for unveiling disease signatures of motoneuron susceptibility: perspectives on the deadly mitochondria, ER stress and calcium triad. Front Cell Neurosci 9:448. https://doi.org/10.3389/fncel.2015.00448
Turner BJ, Atkin JD (2006) ER stress and UPR in familial amyotrophic lateral sclerosis. Curr Mol Med 6(1):79–86
Ilieva EV, Ayala V, Jove M, Dalfo E, Cacabelos D, Povedano M, Bellmunt MJ, Ferrer I et al (2007) Oxidative and endoplasmic reticulum stress interplay in sporadic amyotrophic lateral sclerosis. Brain 130(Pt 12):3111–3123. https://doi.org/10.1093/brain/awm190
Veyrat-Durebex C, Bocca C, Chupin S, Kouassi Nzoughet J, Simard G, Lenaers G, Reynier P, Blasco H (2018) Metabolomics and lipidomics profiling of a combined mitochondrial plus endoplasmic reticulum fraction of human fibroblasts: a robust tool for clinical studies. J Proteome Res 17(1):745–750. https://doi.org/10.1021/acs.jproteome.7b00637
Aviram R, Manella G, Kopelman N, Neufeld-Cohen A, Zwighaft Z, Elimelech M, Adamovich Y, Golik M et al (2016) Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol Cell 62(4):636–648. https://doi.org/10.1016/j.molcel.2016.04.002
Kappler L, Li J, Haring HU, Weigert C, Lehmann R, Xu G, Hoene M (2016) Purity matters: a workflow for the valid high-resolution lipid profiling of mitochondria from cell culture samples. Sci Rep 6:21107. doi:https://doi.org/10.1038/srep21107
Bird SS, Stavrovskaya IG, Gathungu RM, Tousi F, Kristal BS (2015) Qualitative characterization of the rat liver mitochondrial lipidome using all ion fragmentation on an Exactive benchtop Orbitrap MS. Methods Mol Biol 1264:441–452. https://doi.org/10.1007/978-1-4939-2257-4_36
Angelini R, Vitale R, Patil VA, Cocco T, Ludwig B, Greenberg ML, Corcelli A (2012) Lipidomics of intact mitochondria by MALDI-TOF/MS. J Lipid Res 53(7):1417–1425. https://doi.org/10.1194/jlr.D026203
Brooks BR, Miller RG, Swash M, Munsat TL (2000) El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 1(5):293–299
Hutter E, Renner K, Pfister G, Stockl P, Jansen-Durr P, Gnaiger E (2004) Senescence-associated changes in respiration and oxidative phosphorylation in primary human fibroblasts. The Biochem J 380 (Pt 3):919–928. doi:https://doi.org/10.1042/bj20040095
Bocca C, Kane MS, Veyrat-Durebex C, Chupin S, Alban J, Kouassi Nzoughet J, Le Mao M, Chao de la Barca JM et al (2018) The metabolomic bioenergetic signature of Opa1-disrupted mouse embryonic fibroblasts highlights aspartate deficiency. Sci Rep 8(1):11528. https://doi.org/10.1038/s41598-018-29972-9
Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O et al (2007) Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 3(3):211–221. https://doi.org/10.1007/s11306-007-0082-2
Boucret L, Bris C, Seegers V, Goudenege D, Desquiret-Dumas V, Domin-Bernhard M, Ferre-L’Hotellier V, Bouet PE et al (2017) Deep sequencing shows that oocytes are not prone to accumulate mtDNA heteroplasmic mutations during ovarian ageing. Hum Reprod 32(10):2101–2109. https://doi.org/10.1093/humrep/dex268
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. In: Nat Biotechnol, vol 29. vol 1. United States, pp 24–26. doi:https://doi.org/10.1038/nbt.1754
Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC (2013) mtDNA variation and analysis using Mitomap and Mitomaster. Curr Protoc Bioinformatics 44:1.23.21–1.23.26. https://doi.org/10.1002/0471250953.bi0123s44
Clima R, Preste R, Calabrese C, Diroma MA, Santorsola M, Scioscia G, Simone D, Shen L et al (2017) HmtDB 2016: data update, a better performing query system and human mitochondrial DNA haplogroup predictor. Nucleic Acids Res 45(D1):D698–d706. https://doi.org/10.1093/nar/gkw1066
Putz J, Dupuis B, Sissler M, Florentz C (2007) Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA (New York, NY) 13(8):1184–1190. https://doi.org/10.1261/rna.588407
Castellana S, Ronai J, Mazza T (2015) MitImpact: an exhaustive collection of pre-computed pathogenicity predictions of human mitochondrial non-synonymous variants. Hum Mutat 36(2):E2413–E2422. https://doi.org/10.1002/humu.22720
Sonney S, Leipzig J, Lott MT, Zhang S, Procaccio V, Wallace DC, Sondheimer N (2017) Predicting the pathogenicity of novel variants in mitochondrial tRNA with MitoTIP. PLoS Comput Biol 13(12):e1005867. https://doi.org/10.1371/journal.pcbi.1005867
Navarro-Gomez D, Leipzig J, Shen L, Lott M, Stassen AP, Wallace DC, Wiggs JL, Falk MJ, van Oven M, Gai X (2015) Phy-Mer: a novel alignment-free and reference-independent mitochondrial haplogroup classifier. Bioinformatics 31(8):1310–1312. https://doi.org/10.1093/bioinformatics/btu825
Madji Hounoum B, Mavel S, Coque E, Patin F, Vourc’h P, Marouillat S, Nadal-Desbarats L, Emond P et al (2017) Wildtype motoneurons, ALS-linked SOD1 mutation and glutamate profoundly modify astrocyte metabolism and lactate shuttling. Glia 65(4):592–605. https://doi.org/10.1002/glia.23114
Volonte C, Apolloni S, Parisi C, Amadio S (2016) Purinergic contribution to amyotrophic lateral sclerosis. Neuropharmacology 104:180–193. https://doi.org/10.1016/j.neuropharm.2015.10.026
Angermuller S, Islinger M, Volkl A (2009) Peroxisomes and reactive oxygen species, a lasting challenge. Histochem Cell Biol 131(4):459–463. https://doi.org/10.1007/s00418-009-0563-7
Ames BN, Cathcart R, Schwiers E, Hochstein P (1981) Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci U S A 78(11):6858–6862
Sautin YY, Johnson RJ (2008) Uric acid: the oxidant-antioxidant paradox. Nucleosides, Nucleotides Nucleic Acids 27(6):608–619. https://doi.org/10.1080/15257770802138558
Fang P, Li X, Luo JJ, Wang H, Yang XF (2013) A double-edged sword: uric acid and neurological disorders. Brain Disord Ther 2(2):109. https://doi.org/10.4172/2168-975x.1000109
Huang TT, Hao DL, Wu BN, Mao LL, Zhang J (2017) Uric acid demonstrates neuroprotective effect on Parkinson’s disease mice through Nrf2-ARE signaling pathway. Biochem Biophys Res Commun 493(4):1443–1449. https://doi.org/10.1016/j.bbrc.2017.10.004
Bakshi R, Xu Y, Mueller KA, Chen X, Granucci E, Paganoni S, Sadri-Vakili G, Schwarzschild MA (2018) Urate mitigates oxidative stress and motor neuron toxicity of astrocytes derived from ALS-linked SOD1(G93A) mutant mice. Mol Cell Neurosci 92:12–16. https://doi.org/10.1016/j.mcn.2018.06.002
Lazzarino G, Amorini AM, Petzold A, Gasperini C, Ruggieri S, Quartuccio ME, Di Stasio E, Tavazzi B (2017) Serum compounds of energy metabolism impairment are related to disability, disease course and neuroimaging in multiple sclerosis. Mol Neurobiol 54(9):7520–7533. https://doi.org/10.1007/s12035-016-0257-9
Persky AM, Brazeau GA (2001) Clinical pharmacology of the dietary supplement creatine monohydrate. Pharmacol Rev 53(2):161–176
Li H, Tang Z, Chu P, Song Y, Yang Y, Sun B, Niu M, Qaed E et al (2018) Neuroprotective effect of phosphocreatine on oxidative stress and mitochondrial dysfunction induced apoptosis in vitro and in vivo: Involvement of dual PI3K/Akt and Nrf2/HO-1 pathways. Free Radic Biol Med 120:228–238. https://doi.org/10.1016/j.freeradbiomed.2018.03.014
Vallee A, Lecarpentier Y, Guillevin R, Vallee JN (2018) Aerobic glycolysis in amyotrophic lateral sclerosis and Huntington's disease. Rev Neurosci 29(5):547–555. https://doi.org/10.1515/revneuro-2017-0075
Lu W, Su X, Klein MS, Lewis IA, Fiehn O, Rabinowitz JD (2017) Metabolite measurement: pitfalls to avoid and practices to follow. Annu Rev Biochem 86:277–304. https://doi.org/10.1146/annurev-biochem-061516-044952
Farooqui AA, Horrocks LA (2001) Plasmalogens, phospholipase A2, and docosahexaenoic acid turnover in brain tissue. J Mol Neurosci 16(2–3):263–272 discussion 279–284
Peters OM, Ghasemi M, Brown RH Jr (2015) Emerging mechanisms of molecular pathology in ALS. J Clin Invest 125(6):2548. https://doi.org/10.1172/jci82693
Luoma AM, Kuo F, Cakici O, Crowther MN, Denninger AR, Avila RL, Brites P, Kirschner DA (2015) Plasmalogen phospholipids protect internodal myelin from oxidative damage. Free Radic Biol Med 84:296–310. https://doi.org/10.1016/j.freeradbiomed.2015.03.012
Sindelar PJ, Guan Z, Dallner G, Ernster L (1999) The protective role of plasmalogens in iron-induced lipid peroxidation. Free Radic Biol Med 26(3–4):318–324
Blasco H, Veyrat-Durebex C, Bocca C, Patin F, Vourc'h P, Kouassi Nzoughet J, Lenaers G, Andres CR et al (2017) Lipidomics reveals cerebrospinal-fluid signatures of ALS. Sci Rep 7(1):17652. https://doi.org/10.1038/s41598-017-17389-9
Dodge JC, Treleaven CM, Pacheco J, Cooper S, Bao C, Abraham M, Cromwell M, Sardi SP et al (2015) Glycosphingolipids are modulators of disease pathogenesis in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 112(26):8100–8105. https://doi.org/10.1073/pnas.1508767112
Ariga T, Jarvis WD, Yu RK (1998) Role of sphingolipid-mediated cell death in neurodegenerative diseases. J Lipid Res 39(1):1–16
Henriques A, Croixmarie V, Priestman DA, Rosenbohm A, Dirrig-Grosch S, D’Ambra E, Huebecker M, Hussain G et al (2015) Amyotrophic lateral sclerosis and denervation alter sphingolipids and up-regulate glucosylceramide synthase. Hum Mol Genet 24(25):7390–7405. https://doi.org/10.1093/hmg/ddv439
Arima H, Hanada M, Hayasaka T, Masaki N, Omura T, Xu D, Hasegawa T, Togawa D et al (2014) Blockade of IL-6 signaling by MR16-1 inhibits reduction of docosahexaenoic acid-containing phosphatidylcholine levels in a mouse model of spinal cord injury. Neuroscience 269:1–10. https://doi.org/10.1016/j.neuroscience.2014.03.012
Akbar M, Calderon F, Wen Z, Kim HY (2005) Docosahexaenoic acid: a positive modulator of Akt signaling in neuronal survival. Proc Natl Acad Sci U S A 102(31):10858–10863. https://doi.org/10.1073/pnas.0502903102
Hong S, Gronert K, Devchand PR, Moussignac RL, Serhan CN (2003) Novel docosatrienes and 17S-resolvins generated from docosahexaenoic acid in murine brain, human blood, and glial cells. Autacoids in anti-inflammation. J Biol Chem 278(17):14677–14687. https://doi.org/10.1074/jbc.M300218200
Devall M, Mill J, Lunnon K (2014) The mitochondrial epigenome: a role in Alzheimer’s disease? Epigenomics 6(6):665–675. https://doi.org/10.2217/epi.14.50
Hua S, Lu C, Song Y, Li R, Liu X, Quan F, Wang Y, Liu J et al (2012) High levels of mitochondrial heteroplasmy modify the development of ovine-bovine interspecies nuclear transferred embryos. Reprod Fertil Dev 24(3):501–509. https://doi.org/10.1071/rd11091
Stoccoro A, Mosca L, Carnicelli V, Cavallari U, Lunetta C, Marocchi A, Migliore L, Coppede F (2018) Mitochondrial DNA copy number and D-loop region methylation in carriers of amyotrophic lateral sclerosis gene mutations. Epigenomics 10:1431–1443. https://doi.org/10.2217/epi-2018-0072
Acknowledgements
We thank University Hospitals of Angers and Limoges for patients’ recruitment and fibroblasts’ sampling.
Author information
Authors and Affiliations
Contributions
C.V-D and H.B designed and supervised the study, performed statistical analysis and wrote the manuscript; C. Br performed mtDNA experiments and analyzed mtDNA sequence; C.V-D and C.Bo performed metabolomic and lipidomic assays, S.C performed cell cultures and mitochondrial function assessment; P.Cod and B.F recruited ALS patients and performed fibroblast sampling; J.C and P. Cou supervised patients’ recruitment; P.V supervised genetic determination of genes involved in ALS; R.H gave technical and intellectual support and critical advice for English writing; P.Cor, P.V, and C.R.A gave technical and intellectual support; G.L and P.R gave intellectual support and conceptual advice for manuscript writing. All authors offered conceptual advice and comments on the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical Approval and Consent to Participate
All the participants in this current study provided their informed consent for the use of their fibroblasts for research. The ethic committees of the Centre for Human Research of Angers and Limoges Hospitals approved the study and the consent process.
Conflict of Interest
All authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Veyrat-Durebex, C., Bris, C., Codron, P. et al. Metabo-lipidomics of Fibroblasts and Mitochondrial-Endoplasmic Reticulum Extracts from ALS Patients Shows Alterations in Purine, Pyrimidine, Energetic, and Phospholipid Metabolisms. Mol Neurobiol 56, 5780–5791 (2019). https://doi.org/10.1007/s12035-019-1484-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-019-1484-7