Abstract
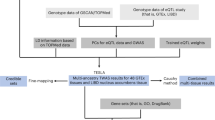
Nicotine has a broad impact on both the central and peripheral nervous systems. Over the past decades, an increasing number of genes potentially involved in nicotine addiction have been identified by different technical approaches. However, the molecular mechanisms underlying nicotine addiction remain largely unknown. Under such situation, prioritizing the candidate genes for further investigation is becoming increasingly important. In this study, we presented a multi-source-based gene prioritization approach for nicotine addiction by utilizing the vast amounts of information generated from for nicotine addiction study during the past years. In this approach, we first collected and curated genes from studies in four categories, i.e., genetic association analysis, genetic linkage analysis, high-throughput gene/protein expression analysis, and literature search of single gene/protein-based studies. Based on these resources, the genes were scored and a weight value was determined for each category. Finally, the genes were ranked by their combined scores, and 220 genes were selected as the prioritized nicotine addiction-related genes. Evaluation suggested the prioritized genes were promising targets for further analysis and replication study.



Similar content being viewed by others
References
Smith SS, Fiore MC (1999) The epidemiology of tobacco use, dependence, and cessation in the United States. Prim Care 26(3):433–461
Murray S (2006) A smouldering epidemic. CMAJ 174(3):309–310
Jha P, Peto R (2014) Global effects of smoking, of quitting, and of taxing tobacco. N Engl J Med 370(1):60–68
Jha P (2009) Avoidable global cancer deaths and total deaths from smoking. Nat Rev Cancer 9(9):655–664
CDC (2007) Cigarette smoking among adults—United States, 2006. Morb Mortal Wkly Rep [serial online] 56(44):1157–1161
CDC (2005b) Annual smoking-attributable mortality, years of potential life lost, and economic costs—United States, 1997--2001. Morb Mortal Wkly Rep [serial online] 54:625–628
Zhang J, Ou JX, Bai CX (2011) Tobacco smoking in China: prevalence, disease burden, challenges and future strategies. Respirology 16(8):1165–1172
Hughes JR, Stead LF, Lancaster T (2007) Antidepressants for smoking cessation. Cochrane Database Syst Rev 1:CD000031
Lerman CE, Schnoll RA, Munafo MR (2007) Genetics and smoking cessation improving outcomes in smokers at risk. Am J Prev Med 33(6 Suppl):S398–S405
Lerman C, Berrettini W (2003) Elucidating the role of genetic factors in smoking behavior and nicotine dependence. Am J Med Genet B Neuropsychiatr Genet 118B(1):48–54
Goode EL et al (2003) Multiple genome-wide analyses of smoking behavior in the Framingham Heart Study. BMC Genet 4(Suppl 1):S102
Osler M et al (2001) Influence of genes and family environment on adult smoking behavior assessed in an adoption study. Genet Epidemiol 21(3):193–200
Tammimaki A et al (2006) Effect of quinpirole on striatal dopamine release and locomotor activity in nicotine-treated mice. Eur J Pharmacol 531(1–3):118–125
Gaddnas H et al (2001) Enhanced motor activity and brain dopamine turnover in mice during long-term nicotine administration in the drinking water. Pharmacol Biochem Behav 70(4):497–503
Benwell ME, Balfour DJ (1992) The effects of acute and repeated nicotine treatment on nucleus accumbens dopamine and locomotor activity. Br J Pharmacol 105(4):849–856
Benwell ME, Balfour DJ (1997) Regional variation in the effects of nicotine on catecholamine overflow in rat brain. Eur J Pharmacol 325(1):13–20
Barik J, Wonnacott S (2006) Indirect modulation by alpha7 nicotinic acetylcholine receptors of noradrenaline release in rat hippocampal slices: interaction with glutamate and GABA systems and effect of nicotine withdrawal. Mol Pharmacol 69(2):618–628
Barik J, Wonnacott S (2009) Molecular and cellular mechanisms of action of nicotine in the CNS. Handb Exp Pharmacol 192:173–207
Kenny PJ, File SE, Rattray M (2001) Nicotine regulates 5-HT(1A) receptor gene expression in the cerebral cortex and dorsal hippocampus. Eur J Neurosci 13(6):1267–1271
Brunzell DH, Russell DS, Picciotto MR (2003) In vivo nicotine treatment regulates mesocorticolimbic CREB and ERK signaling in C57Bl/6J mice. J Neurochem 84(6):1431–1441
Pagliusi SR et al (1996) The reinforcing properties of nicotine are associated with a specific patterning of c-fos expression in the rat brain. Eur J Neurosci 8(11):2247–2256
Nisell M et al (1997) Chronic nicotine enhances basal and nicotine-induced Fos immunoreactivity preferentially in the medial prefrontal cortex of the rat. Neuropsychopharmacology 17(3):151–161
Soderstrom K et al (2007) Nicotine increases FosB expression within a subset of reward- and memory-related brain regions during both peri- and post-adolescence. Psychopharmacology (Berl) 191(4):891–897
Tang K et al (1998) A crucial role for the mitogen-activated protein kinase pathway in nicotinic cholinergic signaling to secretory protein transcription in pheochromocytoma cells. Mol Pharmacol 54(1):59–69
Li MD et al (2004) Time-dependent changes in transcriptional profiles within five rat brain regions in response to nicotine treatment. Brain Res Mol Brain Res 132(2):168–180
Konu O et al (2001) Region-specific transcriptional response to chronic nicotine in rat brain. Brain Res 909(1–2):194–203
Dasgupta P, Chellappan SP (2006) Nicotine-mediated cell proliferation and angiogenesis: new twists to an old story. Cell Cycle 5(20):2324–2328
Dasgupta P et al (2006) Nicotine induces cell proliferation by beta-arrestin-mediated activation of Src and Rb-Raf-1 pathways. J Clin Invest 116(8):2208–2217
Robinson TE, Kolb B (2004) Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology 47(Suppl 1):33–46
Dajas-Bailador F, Wonnacott S (2004) Nicotinic acetylcholine receptors and the regulation of neuronal signalling. Trends Pharmacol Sci 25(6):317–324
Harkness PC, Millar NS (2002) Changes in conformation and subcellular distribution of alpha4beta2 nicotinic acetylcholine receptors revealed by chronic nicotine treatment and expression of subunit chimeras. J Neurosci 22(23):10172–10181
Kane JK et al (2004) Nicotine coregulates multiple pathways involved in protein modification/degradation in rat brain. Brain Res Mol Brain Res 132(2):181–191
Hwang YY, Li MD (2006) Proteins differentially expressed in response to nicotine in five rat brain regions: identification using a 2-DE/MS-based proteomics approach. Proteomics 6(10):3138–3153
Lessov CN et al (2004) Genetics and drug use as a complex phenotype. Subst Use Misuse 39(10–12):1515–1569
Tyndale RF (2003) Genetics of alcohol and tobacco use in humans. Ann Med 35(2):94–121
Hall W, Madden P, Lynskey M (2002) The genetics of tobacco use: methods, findings and policy implications. Tob Control 11(2):119–124
Ho MK, Tyndale RF (2007) Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J 7(2):81–98
Loukola A et al. (2013) Genome-wide association study on detailed profiles of smoking behavior and nicotine dependence in a twin sample. Mol Psychiatry
Thorgeirsson TE et al (2010) Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 42(5):448–453
Uhl GR et al (2008) Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry 65(6):683–693
Wang J et al. (2007) Strain- and region-specific gene expression profiles in mouse brain in response to chronic nicotine treatment. Genes Brain Behav
Wang J et al (2009) Significant modulation of mitochondrial electron transport system by nicotine in various rat brain regions. Mitochondrion 9(3):186–195
Wang J, Yuan W, Li MD (2011) Genes and pathways co-associated with the exposure to multiple drugs of abuse, including alcohol, amphetamine/methamphetamine, cocaine, marijuana, morphine, and/or nicotine: a review of proteomics analyses. Mol Neurobiol 44(3):269–286
Blalock EM et al (2005) Harnessing the power of gene microarrays for the study of brain aging and Alzheimer's disease: statistical reliability and functional correlation. Ageing Res Rev 4(4):481–512
Pearson TA, Manolio TA (2008) How to interpret a genome-wide association study. JAMA 299(11):1335–1344
Wang J, Li MD (2010) Common and unique biological pathways associated with smoking initiation/progression, nicotine dependence, and smoking cessation. Neuropsychopharmacology 35(3):702–719
Sullivan PF et al (2004) Candidate genes for nicotine dependence via linkage, epistasis, and bioinformatics. Am J Med Genet B Neuropsychiatr Genet 126B(1):23–36
Bierut LJ et al (2007) Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet 16(1):24–35
Saccone SF et al (2007) Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 16(1):36–49
Uhl GR et al (2007) Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet 8:10
Han S et al (2010) Meta-analysis of 15 genome-wide linkage scans of smoking behavior. Biol Psychiatry 67(1):12–19
Roberts PM (2006) Mining literature for systems biology. Brief Bioinform 7(4):399–406
Li CY, Mao X, Wei L (2008) Genes and (common) pathways underlying drug addiction. PLoS Comput Biol 4(1):e2
Li MD, Burmeister M (2009) New insights into the genetics of addiction. Nat Rev Genet 10(4):225–231
Dekkers A, Arats E (1991) Global optimization and simulated annealing. Mathmatical Programing 50(1-3):367–393
Corana A et al (1987) Minimizing multimodal functions of continuous variables with the ‘simulated annealing’ algorithm. ACM Trans Math Softw 13(3):262–280
Locatelli M (2000) Simulated annealing algorithms for continuous global optimization. J Optim Theory Appl 104(1):121–133
Chen J et al (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37(Web Server issue):W305–W311
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57(1):289–300
Sun J, Zhao Z (2010) Functional features, biological pathways, and protein interaction networks of addiction-related genes. Chem Biodivers 7(5):1153–1162
Zhu J, Reith ME (2008) Role of the dopamine transporter in the action of psychostimulants, nicotine, and other drugs of abuse. CNS Neurol Disord Drug Targets 7(5):393–409
Sulzer D (2011) How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron 69(4):628–649
Konneker T et al (2008) A searchable database of genetic evidence for psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet 147B(6):671–675
Sahoo SS et al (2008) An ontology-driven semantic mashup of gene and biological pathway information: application to the domain of nicotine dependence. J Biomed Inform 41(5):752–765
Saccone SF et al (2008) Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics 24(16):1805–1811
Lewinger JP et al (2007) Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet Epidemiol 31(8):871–882
Acknowledgments
This work was supported in part by grants from National Natural Science Foundation of China (Grant No. 31271411) and Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry of China. We are grateful to Prof. Ming D Li of University of Virginia for his help on this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xinhua Liu and Meng Liu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, X., Liu, M., Li, X. et al. Prioritizing Genes Related to Nicotine Addiction Via a Multi-source-Based Approach. Mol Neurobiol 52, 442–455 (2015). https://doi.org/10.1007/s12035-014-8874-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-8874-7