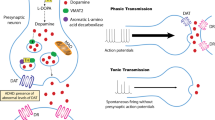
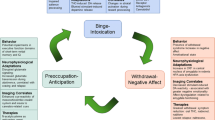
Abstract
Drug addiction is a chronic neuronal disease. In recent years, proteomics technology has been widely used to assess the protein expression in the brain tissues of both animals and humans exposed to addictive drugs. Through this approach, a large number of proteins potentially involved in the etiology of drug addictions have been identified, which provide a valuable resource to study protein function, biochemical pathways, and networks related to the molecular mechanisms underlying drug dependence. In this article, we summarize the recent application of proteomics to profiling protein expression patterns in animal or human brain tissues after the administration of alcohol, amphetamine/methamphetamine, cocaine, marijuana, morphine/heroin/butorphanol, or nicotine. From available reports, we compiled a list of 497 proteins associated with exposure to one or more addictive drugs, with 160 being related to exposure to at least two abused drugs. A number of biochemical pathways and biological processes appear to be enriched among these proteins, including synaptic transmission and signaling pathways related to neuronal functions. The data included in this work provide a summary and extension of the proteomics studies on drug addiction. Furthermore, the proteins and biological processes highlighted here may provide valuable insight into the cellular activities and biological processes in neurons in the development of drug addiction.


Similar content being viewed by others
References
Potenza MN, Sofuoglu M, Carroll KM, Rounsaville BJ (2011) Neuroscience of behavioral and pharmacological treatments for addictions. Neuron 69:695–712
Nestler EJ (2004) Molecular mechanisms of drug addiction. Neuropharmacology 47(Suppl 1):24–32
Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, Nestler EJ (2010) The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci 33:267–276
Tan KR, Rudolph U, Luscher C (2011) Hooked on benzodiazepines: GABAA receptor subtypes and addiction. Trends Neurosci 34:188–197
Luscher C, Malenka RC (2011) Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron 69:650–663
Wong CC, Mill J, Fernandes C (2011) Drugs and addiction: an introduction to epigenetics. Addiction 106:480–489
Renthal W, Nestler EJ (2009) Chromatin regulation in drug addiction and depression. Dialogues Clin Neurosci 11:257–268
Nestler EJ (2001) Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci 2:119–128
Larsson A, Engel JA (2004) Neurochemical and behavioral studies on ethanol and nicotine interactions. Neurosci Biobehav Rev 27:713–720
Hemby SE (2010) Cocainomics: new insights into the molecular basis of cocaine addiction. J Neuroimmune Pharmacol 5:70–82
Hwang YY, Li MD (2006) Proteins differentially expressed in response to nicotine in five rat brain regions: identification using a 2-DE/MS-based proteomics approach. Proteomics 6:3138–3153
Bierczynska-Krzysik A, Pradeep John JP, Silberring J, Kotlinska J, Dylag T, Cabatic M, Lubec G (2006) Proteomic analysis of rat cerebral cortex, hippocampus and striatum after exposure to morphine. Int J Mol Med 18:775–784
Freeman WM, Hemby SE (2004) Proteomics for protein expression profiling in neuroscience. Neurochem Res 29:1065–1081
Tannu N, Mash DC, Hemby SE (2007) Cytosolic proteomic alterations in the nucleus accumbens of cocaine overdose victims. Mol Psychiatry 12:55–73
Witzmann FA, Strother WN (2004) Proteomics and alcoholism. Int Rev Neurobiol 61:189–214
Lull ME, Freeman WM, VanGuilder HD, Vrana KE (2010) The use of neuroproteomics in drug abuse research. Drug Alcohol Depend 107:11–22
Li MD, Wang J (2007) Neuroproteomics and its applications in research on nicotine and other drugs of abuse. Proteomics Clin Appl 1:1406–1427
Witzmann FA, Li J, Strother WN, McBride WJ, Hunter L, Crabb DW, Lumeng L, Li TK (2003) Innate differences in protein expression in the nucleus accumbens and hippocampus of inbred alcohol-preferring and -nonpreferring rats. Proteomics 3:1335–1344
Bell RL, Kimpel MW, Rodd ZA, Strother WN, Bai F, Peper CL, Mayfield RD, Lumeng L, Crabb DW, McBride WJ et al (2006) Protein expression changes in the nucleus accumbens and amygdala of inbred alcohol-preferring rats given either continuous or scheduled access to ethanol. Alcohol 40:3–17
McBride WJ, Schultz JA, Kimpel MW, McClintick JN, Wang M, You J, Rodd ZA (2009) Differential effects of ethanol in the nucleus accumbens shell of alcohol-preferring (P), alcohol-non-preferring (NP) and Wistar rats: a proteomics study. Pharmacol Biochem Behav 92:304–313
Damodaran S, Dlugos CA, Wood TD, Rabin RA (2006) Effects of chronic ethanol administration on brain protein levels: a proteomic investigation using 2-D DIGE system.Eur J Pharmacol 547:75–82
Park B, Jeong SK, Lee WS, Seong JK, Paik YK (2004) A simple pattern classification method for alcohol-responsive proteins that are differentially expressed in mouse brain. Proteomics 4:3369–3375
Sari Y, Zhang M, Mechref Y (2010) Differential expression of proteins in fetal brains of alcohol-treated prenatally C57BL/6 mice: a proteomic investigation. Electrophoresis 31:483–496
Lewohl JM, Van Dyk DD, Craft GE, Innes DJ, Mayfield RD, Cobon G, Harris RA, Dodd PR (2004) The application of proteomics to the human alcoholic brain. Ann N Y Acad Sci 1025:14–26
Alexander-Kaufman K, James G, Sheedy D, Harper C, Matsumoto I (2006) Differential protein expression in the prefrontal white matter of human alcoholics: a proteomics study. Mol Psychiatry 11:56–65
Alexander-Kaufman K, Cordwell S, Harper C, Matsumoto I (2007) A proteome analysis of the dorsolateral prefrontal cortex in human alcoholic patients. Proteomics Clin Appl 1:62–72
Alexander-Kaufman K, Harper C, Wilce P, Matsumoto I (2007) Cerebellar vermis proteome of chronic alcoholic individuals. Alcohol Clin Exp Res 31:1286–1296
Matsumoto I (2009) Proteomics approach in the study of the pathophysiology of alcohol-related brain damage. Alcohol Alcohol 44:171–176
Kashem MA, Etages HD, Kopitar-Jerala N, McGregor IS, Matsumoto I (2009) Differential protein expression in the corpus callosum (body) of human alcoholic brain. J Neurochem 110:486–495
Kashem MA, Harper C, Matsumoto I (2008) Differential protein expression in the corpus callosum (genu) of human alcoholics. Neurochem Int 53:1–11
Kashem MA, James G, Harper C, Wilce P, Matsumoto I (2007) Differential protein expression in the corpus callosum (splenium) of human alcoholics: a proteomics study. Neurochem Int 50:450–459
Matsuda-Matsumoto H, Iwazaki T, Kashem MA, Harper C, Matsumoto I (2007) Differential protein expression profiles in the hippocampus of human alcoholics. Neurochem Int 51:370–376
Freeman WM, Brebner K, Amara SG, Reed MS, Pohl J, Phillips AG (2005) Distinct proteomic profiles of amphetamine self-administration transitional states. Pharmacogenomics J 5:203–214
Iwazaki T, McGregor IS, Matsumoto I (2006) Protein expression profile in the striatum of acute methamphetamine-treated rats. Brain Res 1097:19–25
Iwazaki T, McGregor IS, Matsumoto I (2007) Protein expression profile in the striatum of rats with methamphetamine-induced behavioral sensitization. Proteomics 7:1131–1139
Iwazaki T, McGregor IS, Matsumoto I (2008) Protein expression profile in the amygdala of rats with methamphetamine-induced behavioral sensitization. Neurosci Lett 435:113–119
Kobeissy FH, Warren MW, Ottens AK, Sadasivan S, Zhang Z, Gold MS, Wang KK (2008) Psychoproteomic analysis of rat cortex following acute methamphetamine exposure. J Proteome Res 7:1971–1983
Faure JJ, Hattingh SM, Stein DJ, Daniels WM (2009) Proteomic analysis reveals differentially expressed proteins in the rat frontal cortex after methamphetamine treatment. Metab Brain Dis 24:685–700
Li X, Wang H, Qiu P, Luo H (2008) Proteomic profiling of proteins associated with methamphetamine-induced neurotoxicity in different regions of rat brain. Neurochem Int 52:256–264
Yang MH, Kim S, Jung MS, Shim JH, Ryu NK, Yook YJ, Jang CG, Bahk YY, Kim KW, Park JH (2008) Proteomic analysis of methamphetamine-induced reinforcement processes within the mesolimbic dopamine system. Addict Biol 13:287–294
Lull ME, Freeman WM, Vrana KE, Mash DC (2008) Correlating human and animal studies of cocaine abuse and gene expression. Ann N Y Acad Sci 1141:58–75
Hemby SE (2006) Assessment of genome and proteome profiles in cocaine abuse. Prog Brain Res 158:173–195
Tannu NS, Howell LL, Hemby SE (2010) Integrative proteomic analysis of the nucleus accumbens in rhesus monkeys following cocaine self-administration. Mol Psychiatry 15:185–203
del Castillo C, Morales L, Alguacil LF, Salas E, Garrido E, Alonso E, Perez-Garcia C (2009) Proteomic analysis of the nucleus accumbens of rats with different vulnerability to cocaine addiction. Neuropharmacology 57:41–48
Reynolds JL, Mahajan SD, Bindukumar B, Sykes D, Schwartz SA, Nair MP (2006) Proteomic analysis of the effects of cocaine on the enhancement of HIV-1 replication in normal human astrocytes (NHA). Brain Res 1123:226–236
Bindukumar B, Mahajan SD, Reynolds JL, Hu Z, Sykes DE, Aalinkeel R, Schwartz SA (2008) Genomic and proteomic analysis of the effects of cannabinoids on normal human astrocytes. Brain Res 1191:1–11
Quinn HR, Matsumoto I, Callaghan PD, Long LE, Arnold JC, Gunasekaran N, Thompson MR, Dawson B, Mallet PE, Kashem MA et al (2008) Adolescent rats find repeated Delta(9)-THC less aversive than adult rats but display greater residual cognitive deficits and changes in hippocampal protein expression following exposure. Neuropsychopharmacology 33:1113–1126
Rubino T, Realini N, Braida D, Alberio T, Capurro V, Vigano D, Guidali C, Sala M, Fasano M, Parolaro D (2009) The depressive phenotype induced in adult female rats by adolescent exposure to THC is associated with cognitive impairment and altered neuroplasticity in the prefrontal cortex. Neurotox Res 15:291–302
Colombo G, Rusconi F, Rubino T, Cattaneo A, Martegani E, Parolaro D, Bachi A, Zippel R (2009) Transcriptomic and proteomic analyses of mouse cerebellum reveals alterations in RasGRF1 expression following in vivo chronic treatment with delta 9-tetrahydrocannabinol. J Mol Neurosci 37:111–122
Li KW, Jimenez CR, van der Schors RC, Hornshaw MP, Schoffelmeer AN, Smit AB (2006) Intermittent administration of morphine alters protein expression in rat nucleus accumbens. Proteomics 6:2003–2008
Kim SY, Chudapongse N, Lee SM, Levin MC, Oh JT, Park HJ, Ho IK (2005) Proteomic analysis of phosphotyrosyl proteins in morphine-dependent rat brains. Brain Res Mol Brain Res 133:58–70
Bodzon-Kulakowska A, Suder P, Mak P, Bierczynska-Krzysik A, Lubec G, Walczak B, Kotlinska J, Silberring J (2009) Proteomic analysis of striatal neuronal cell cultures after morphine administration. J Sep Sci 32:1200–1210
Bierczynska-Krzysik A, Bonar E, Drabik A, Noga M, Suder P, Dylag T, Dubin A, Kotlinska J, Silberring J (2006) Rat brain proteome in morphine dependence. Neurochem Int 49:401–406
Shui HA, Ho ST, Wang JJ, Wu CC, Lin CH, Tao YX, Liaw WJ (2007) Proteomic analysis of spinal protein expression in rats exposed to repeated intrathecal morphine injection. Proteomics 7:796–803
Li Q, Zhao X, Zhong LJ, Yang HY, Wang Q, Pu XP (2009) Effects of chronic morphine treatment on protein expression in rat dorsal root ganglia. Eur J Pharmacol 612:21–28
Neasta J, Uttenweiler-Joseph S, Chaoui K, Monsarrat B, Meunier JC, Mouledous L (2006) Effect of long-term exposure of SH-SY5Y cells to morphine: a whole cell proteomic analysis. Proteome Sci 4:23
Suder P, Bodzon-Kulakowska A, Mak P, Bierczynska-Krzysik A, Daszykowski M, Walczak B, Lubec G, Kotlinska JH, Silberring J (2009) The proteomic analysis of primary cortical astrocyte cell culture after morphine administration. J Proteome Res 8:4633–4640
Moron JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R, Devi LA (2007) Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: a proteomics study focusing on endocytic proteins. Mol Cell Proteomics 6:29–42
Van den Oever MC, Lubbers BR, Goriounova NA, Li KW, Van der Schors RC, Loos M, Riga D, Wiskerke J, Binnekade R, Stegeman M et al (2010) Extracellular matrix plasticity and GABAergic inhibition of prefrontal cortex pyramidal cells facilitates relapse to heroin seeking. Neuropsychopharmacology 35:2120–2133
Prokai L, Zharikova AD, Stevens SM Jr (2005) Effect of chronic morphine exposure on the synaptic plasma-membrane subproteome of rats: a quantitative protein profiling study based on isotope-coded affinity tags and liquid chromatography/mass spectrometry. J Mass Spectrom 40:169–175
Yang L, Sun ZS, Zhu YP (2007) Proteomic analysis of rat prefrontal cortex in three phases of morphine-induced conditioned place preference. J Proteome Res 6:2239–2247
Bodzon-Kulakowska A, Kulakowski K, Drabik A, Moszczynski A, Silberring J, Suder P (2011) Morphinome—a meta-analysis applied to proteomics studies in morphine dependence. Proteomics 11:5–21
Bodzon-Kulakowska A, Bierczynska-Krzysik A, Drabik A, Noga M, Kraj A, Suder P, Silberring J (2005) Morphinome—proteome of the nervous system after morphine treatment. Amino Acids 28:13–19
Kim SY, Chudapongse N, Lee SM, Levin MC, Oh JT, Park HJ, Ho IK (2004) Proteomic analysis of phosphotyrosyl proteins in the rat brain: effect of butorphanol dependence. J Neurosci Res 77:867–877
Yeom M, Shim I, Lee HJ, Hahm DH (2005) Proteomic analysis of nicotine-associated protein expression in the striatum of repeated nicotine-treated rats. Biochem Biophys Res Commun 326:321–328
Benowitz NL (1996) Pharmacology of nicotine: addiction and therapeutics. Annu Rev Pharmacol Toxicol 36:597–613
Piubelli C, Cecconi D, Astner H, Caldara F, Tessari M, Carboni L, Hamdan M, Righetti PG, Domenici E (2005) Proteomic changes in rat serum, polymorphonuclear and mononuclear leukocytes after chronic nicotine administration. Proteomics 5:1382–1394
Cecconi D, Tessari M, Wille DR, Zoli M, Domenici E, Righetti PG, Carboni L (2008) Serum proteomic analysis during nicotine self-administration, extinction nd relapse in rats. Electrophoresis 29:1525–1533
Jahn R, Scheller RH (2006) SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol 7:631–643
Ungermann C, Langosch D (2005) Functions of SNAREs in intracellular membrane fusion and lipid bilayer mixing. J Cell Sci 118:3819–3828
Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116:153–166
Schweizer FE, Ryan TA (2006) The synaptic vesicle: cycle of exocytosis and endocytosis. Curr Opin Neurobiol 16:298–304
Kennedy MJ, Ehlers MD (2006) Organelles and trafficking machinery for postsynaptic plasticity. Annu Rev Neurosci 29:325–362
Clayton EL, Cousin MA (2009) The molecular physiology of activity-dependent bulk endocytosis of synaptic vesicles. J Neurochem 111:901–914
Kennedy MJ, Ehlers MD (2011) Mechanisms and function of dendritic exocytosis. Neuron 69:856–875
Xu Q, Li MD (2011) Nicotine modulates expression of dynamin 1 in rat brain and SH-SY5Y cells. Neurosci Lett 489:168–171
Huang W, Li MD (2009) Nicotine modulates expression of miR-140*, which targets the 3′-untranslated region of dynamin 1 gene (Dnm1). Int J Neuropsychopharmacol 12:537–546
Garcia-Fuster MJ, Ferrer-Alcon M, Miralles A, Garcia-Sevilla JA (2003) Modulation of Fas receptor proteins and dynamin during opiate addiction and induction of opiate withdrawal in rat brain. Naunyn Schmiedebergs Arch Pharmacol 368:421–431
Saito M, Smiley J, Toth R, Vadasz C (2002) Microarray analysis of gene expression in rat hippocampus after chronic ethanol treatment. Neurochem Res 27:1221–1229
Sontag JM, Fykse EM, Ushkaryov Y, Liu JP, Robinson PJ, Sudhof TC (1994) Differential expression and regulation of multiple dynamins. J Biol Chem 269:4547–4554
Shupliakov O, Low P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L (1997) Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science 276:259–263
Takei K, Mundigl O, Daniell L, De Camilli P (1996) The synaptic vesicle cycle: a single vesicle budding step involving clathrin and dynamin. J Cell Biol 133:1237–1250
Robinson PJ, Liu JP, Powell KA, Fykse EM, Sudhof TC (1994) Phosphorylation of dynamin I and synaptic-vesicle recycling. Trends Neurosci 17:348–353
Farr CD, Gafken PR, Norbeck AD, Doneanu CE, Stapels MD, Barofsky DF, Minami M, Saugstad JA (2004) Proteomic analysis of native metabotropic glutamate receptor 5 protein complexes reveals novel molecular constituents. J Neurochem 91:438–450
Kranenburg O, Verlaan I, Moolenaar WH (1999) Dynamin is required for the activation of mitogen-activated protein (MAP) kinase by MAP kinase kinase. J Biol Chem 274:35301–35304
Whistler JL, von Zastrow M (1999) Dissociation of functional roles of dynamin in receptor-mediated endocytosis and mitogenic signal transduction. J Biol Chem 274:24575–24578
Bennett MK, Calakos N, Scheller RH (1992) Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 257:255–259
Smirnova T, Stinnakre J, Mallet J (1993) Characterization of a presynaptic glutamate receptor. Science 262:430–433
Pevsner J, Hsu SC, Scheller RH (1994) n-Sec1: a neural-specific syntaxin-binding protein. Proc Natl Acad Sci U S A 91:1445–1449
Greengard P, Valtorta F, Czernik AJ, Benfenati F (1993) Synaptic vesicle phosphoproteins and regulation of synaptic function. Science 259:780–785
Wang Y, Tang BL (2006) SNAREs in neurons—beyond synaptic vesicle exocytosis (review). Mol Membr Biol 23:377–384
Osei YD, Churchich JE (1995) Screening and sequence determination of a cDNA encoding the human brain 4-aminobutyrate aminotransferase. Gene 155:185–187
De Biase D, Barra D, Simmaco M, John RA, Bossa F (1995) Primary structure and tissue distribution of human 4-aminobutyrate aminotransferase. Eur J Biochem 227:476–480
Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP (2005) Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet 76:950–966
Clancy KP, Berger R, Cox M, Bleskan J, Walton KA, Hart I, Patterson D (1996) Localization of the l-glutamine synthetase gene to chromosome 1q23. Genomics 38:418–420
Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM (2001) Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell 104:675–686
Mansuy IM, Mayford M, Jacob B, Kandel ER, Bach ME (1998) Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell 92:39–49
Mulkey RM, Endo S, Shenolikar S, Malenka RC (1994) Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature 369:486–488
Jin Z, Gao F, Flagg T, Deng X (2004) Nicotine induces multi-site phosphorylation of Bad in association with suppression of apoptosis. J Biol Chem 279:23837–23844
Exley R, Moroni M, Sasdelli F, Houlihan LM, Lukas RJ, Sher E, Zwart R, Bermudez I (2006) Chaperone protein 14-3-3 and protein kinase A increase the relative abundance of low agonist sensitivity human alpha 4 beta 2 nicotinic acetylcholine receptors in Xenopus oocytes. J Neurochem 98:876–885
Jeanclos EM, Lin L, Treuil MW, Rao J, DeCoster MA, Anand R (2001) The chaperone protein 14-3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization. J Biol Chem 276:28281–28290
Rajalingam K, Rudel T (2005) Ras–Raf signaling needs prohibitin. Cell Cycle 4:1503–1505
Fu H, Xia K, Pallas DC, Cui C, Conroy K, Narsimhan RP, Mamon H, Collier RJ, Roberts TM (1994) Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science 266:126–129
Hermeking H, Benzinger A (2006) 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol 16:183–192
Nestler EJ (2005) Is there a common molecular pathway for addiction? Nat Neurosci 8:1445–1449
Kauer JA, Malenka RC (2007) Synaptic plasticity and addiction. Nat Rev Neurosci 8:844–858
Wang J, Li MD (2010) Common and unique biological pathways associated with smoking initiation/progression, nicotine dependence, and smoking cessation. Neuropsychopharmacology 35:702–719
Mukherjee S, Das SK, Vaidyanathan K, Vasudevan DM (2008) Consequences of alcohol consumption on neurotransmitters—an overview. Curr Neurovasc Res 5:266–272
Nestler EJ (1997) Molecular mechanisms of opiate and cocaine addiction. Curr Opin Neurobiol 7:713–719
Liu Z, Tearle AW, Nai Q, Berg DK (2005) Rapid activity-driven SNARE-dependent trafficking of nicotinic receptors on somatic spines. J Neurosci 25:1159–1168
Roy S, Wyse B, Hancock JF (2002) H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol Cell Biol 22:5128–5140
Ferre S, Lluis C, Justinova Z, Quiroz C, Orru M, Navarro G, Canela EI, Franco R, Goldberg SR (2010) Adenosine-cannabinoid receptor interactions. Implications for striatal function. Br J Pharmacol 160:443–453
Contet C, Kieffer BL, Befort K (2004) Mu opioid receptor: a gateway to drug addiction. Curr Opin Neurobiol 14:370–378
Changeux JP (2010) Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nat Rev Neurosci 11:389–401
Huber LA (2003) Is proteomics heading in the wrong direction? Nat Rev Mol Cell Biol 4:74–80
Garbis S, Lubec G, Fountoulakis M (2005) Limitations of current proteomics technologies. J Chromatogr A 1077:1–18
Hargreaves GA, Quinn H, Kashem MA, Matsumoto I, McGregor IS (2009) Proteomic analysis demonstrates adolescent vulnerability to lasting hippocampal changes following chronic alcohol consumption. Alcohol Clin Exp Res 33:86–94
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc, Series B 57:289–300
Acknowledgments
The preparation of this article was in part supported by National Institutes of Health grants DA-12844 and DA-13783. We thank Dr. David L. Bronson for his excellent editing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1
(DOC 775 kb)
Rights and permissions
About this article
Cite this article
Wang, J., Yuan, W. & Li, M.D. Genes and Pathways Co-associated with the Exposure to Multiple Drugs of Abuse, Including Alcohol, Amphetamine/Methamphetamine, Cocaine, Marijuana, Morphine, and/or Nicotine: a Review of Proteomics Analyses. Mol Neurobiol 44, 269–286 (2011). https://doi.org/10.1007/s12035-011-8202-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-011-8202-4