Abstract
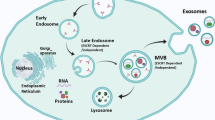
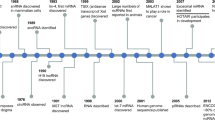
Exosomes are extra-cellular vesicles that are < 150 nm that is formed by invagination of the plasma membrane and are released as vesicles. These contain proteins, RNA, and DNA as their cargo. In recent times, the non-coding RNA (ncRNA) present within exosomes has been studied extensively in the context of sorting, localization, and their potential as biomarkers. For example, miR-1246, miR-1290, miR-21, and miR-23a are exosomal biomarkers of cancer, and YBX1 (Y-Box Binding Protein 1) is attributed to exosomal RNA sorting. Transfer RNA-derived fragments are a class of small ncRNAs that were discovered in 2009. They are classified as tRFs (tRNA-derived fragments) and tsRNAs (tRNA halves). Interestingly, these tRNA-derived ncRNAs are emerging as biomarkers in various diseases, and these are found in exosomes. To date, the literature has covered only the biomarker potential of plasma/serum tRNA-derived ncRNAs. Hence, in the current review, we discuss the exosomal tRNA-derived fragments that are clinically relevant in pathological conditions.



Similar content being viewed by others
Data Availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Abbreviations
- ncRNA:
-
Non-coding RNA
- tRFs:
-
tRNA-derived fragments
- tsRNAs:
-
tRNA halves
- YBX1:
-
Y box-binding protein 1
- CKD:
-
Chronic kidney disease
- SE:
-
Seminal exosomes
- SONFH:
-
Steroid-induced osteonecrosis of the femoral head
- AD:
-
Atopic dermatitis
References
Gurunathan, S., Kang, M. H., Jeyaraj, M., Qasim, M., & Kim, J. H. (2019). Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. https://doi.org/10.3390/cells8040307
Wolf, P. (1967). The nature and significance of platelet products in human plasma. British Journal of Haematology, 13, 269–288. https://doi.org/10.1111/j.1365-2141.1967.tb08741.x
Doyle, L. M., & Wang, M. Z. (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. https://doi.org/10.3390/cells8070727
Baietti, M. F., Zhang, Z., Mortier, E., Melchior, A., Degeest, G., Geeraerts, A., Ivarsson, Y., Depoortere, F., Coomans, C., Vermeiren, E., Zimmermann, P., & David, G. (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nature Cell Biology, 14, 677–685. https://doi.org/10.1038/ncb2502
Gruenberg, J., & van der Goot, F. G. (2006). Mechanisms of pathogen entry through the endosomal compartments. Nature Reviews Molecular Cell Biology, 7, 495–504. https://doi.org/10.1038/nrm1959
Henne, W. M., Stenmark, H., & Emr, S. D. (2013). Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harbor Perspectives in Biology. https://doi.org/10.1101/cshperspect.a016766
Ostrowski, M., Carmo, N. B., Krumeich, S., Fanget, I., Raposo, G., Savina, A., Moita, C. F., Schauer, K., Hume, A. N., Freitas, R. P., Goud, B., Benaroch, P., Hacohen, N., Fukuda, M., Desnos, C., Seabra, M. C., Darchen, F., Amigorena, S., Moita, L. F., & Thery, C. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature Cell Biology, 12, 19–30; sup pp 1–13. https://doi.org/10.1038/ncb2000
Razi, M., & Futter, C. E. (2006). Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Molecular Biology of the Cell, 17, 3469–3483. https://doi.org/10.1091/mbc.e05-11-1054
Joyce, D. P., Kerin, M. J., & Dwyer, R. M. (2016). Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. International Journal of Cancer, 139, 1443–1448. https://doi.org/10.1002/ijc.30179
Zhang, J., Li, S., Li, L., Li, M., Guo, C., Yao, J., & Mi, S. (2015). Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics, Proteomics & Bioinformatics, 13, 17–24. https://doi.org/10.1016/j.gpb.2015.02.001
Janowska-Wieczorek, A., Wysoczynski, M., Kijowski, J., Marquez-Curtis, L., Machalinski, B., Ratajczak, J., & Ratajczak, M. Z. (2005). Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. International Journal of Cancer, 113, 752–760. https://doi.org/10.1002/ijc.20657
Zhu, L., Sun, H. T., Wang, S., Huang, S. L., Zheng, Y., Wang, C. Q., Hu, B. Y., Qin, W., Zou, T. T., Fu, Y., Shen, X. T., Zhu, W. W., Geng, Y., Lu, L., Jia, H. L., Qin, L. X., & Dong, Q. Z. (2020). Isolation and characterization of exosomes for cancer research. Journal of Hematology & Oncology, 13, 152. https://doi.org/10.1186/s13045-020-00987-y
Greening, D. W., Xu, R., Ji, H., Tauro, B. J., & Simpson, R. J. (2015). A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods in Molecular Biology, 1295, 179–209. https://doi.org/10.1007/978-1-4939-2550-6_15
Caradec, J., Kharmate, G., Hosseini-Beheshti, E., Adomat, H., Gleave, M., & Guns, E. (2014). Reproducibility and efficiency of serum-derived exosome extraction methods. Clinical Biochemistry, 47, 1286–1292. https://doi.org/10.1016/j.clinbiochem.2014.06.011
Oksvold, M. P., Neurauter, A., & Pedersen, K. W. (2015). Magnetic bead-based isolation of exosomes. Methods in Molecular Biology, 1218, 465–481. https://doi.org/10.1007/978-1-4939-1538-5_27
Weng, Y., Sui, Z., Shan, Y., Hu, Y., Chen, Y., Zhang, L., & Zhang, Y. (2016). Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. The Analyst, 141, 4640–4646. https://doi.org/10.1039/c6an00892e
Arroyo, J. D., Chevillet, J. R., Kroh, E. M., Ruf, I. K., Pritchard, C. C., Gibson, D. F., Mitchell, P. S., Bennett, C. F., Pogosova-Agadjanyan, E. L., Stirewalt, D. L., Tait, J. F., & Tewari, M. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A, 108, 5003–5008. https://doi.org/10.1073/pnas.1019055108
Tabet, F., Vickers, K. C., Cuesta Torres, L. F., Wiese, C. B., Shoucri, B. M., Lambert, G., Catherinet, C., Prado-Lourenco, L., Levin, M. G., Thacker, S., Sethupathy, P., Barter, P. J., Remaley, A. T., & Rye, K. A. (2014). HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nature Communications, 5, 3292. https://doi.org/10.1038/ncomms4292
Melo, S. A., Sugimoto, H., O’Connell, J. T., Kato, N., Villanueva, A., Vidal, A., Qiu, L., Vitkin, E., Perelman, L. T., Melo, C. A., Lucci, A., Ivan, C., Calin, G. A., & Kalluri, R. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell, 26, 707–721. https://doi.org/10.1016/j.ccell.2014.09.005
Fabbri, M., Paone, A., Calore, F., Galli, R., Gaudio, E., Santhanam, R., Lovat, F., Fadda, P., Mao, C., Nuovo, G. J., Zanesi, N., Crawford, M., Ozer, G. H., Wernicke, D., Alder, H., Caligiuri, M. A., Nana-Sinkam, P., Perrotti, D., & Croce, C. M. (2012). MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A, 109, E2110–E2116. https://doi.org/10.1073/pnas.1209414109
Ogata-Kawata, H., Izumiya, M., Kurioka, D., Honma, Y., Yamada, Y., Furuta, K., Gunji, T., Ohta, H., Okamoto, H., Sonoda, H., Watanabe, M., Nakagama, H., Yokota, J., Kohno, T., & Tsuchiya, N. (2014). Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE, 9, e92921. https://doi.org/10.1371/journal.pone.0092921
Wang, J. M., Ju, B. H., Pan, C. J., Gu, Y., Li, M. Q., Sun, L., Xu, Y. Y., & Yin, L. R. (2017). MiR-214 inhibits cell migration, invasion and promotes the drug sensitivity in human cervical cancer by targeting FOXM1. Am J Transl Res, 9, 3541–3557.
Honda, S., Kawamura, T., Loher, P., Morichika, K., Rigoutsos, I., & Kirino, Y. (2017). The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Research, 45, 9108–9120. https://doi.org/10.1093/nar/gkx537
Sun, C., Fu, Z., Wang, S., Li, J., Li, Y., Zhang, Y., Yang, F., Chu, J., Wu, H., Huang, X., Li, W., & Yin, Y. (2018). Roles of tRNA-derived fragments in human cancers. Cancer Letters, 414, 16–25. https://doi.org/10.1016/j.canlet.2017.10.031
Goodarzi, H., Liu, X., Nguyen, H. C., Zhang, S., Fish, L., & Tavazoie, S. F. (2015). Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell, 161, 790–802. https://doi.org/10.1016/j.cell.2015.02.053
Kuscu, C., Kumar, P., Kiran, M., Su, Z., Malik, A., & Dutta, A. (2018). tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA, 24, 1093–1105. https://doi.org/10.1261/rna.066126.118
Guan, L., Karaiskos, S., & Grigoriev, A. (2020). Inferring targeting modes of Argonaute-loaded tRNA fragments. RNA Biology, 17, 1070–1080. https://doi.org/10.1080/15476286.2019.1676633
Choi, E. J., Ren, J., Zhang, K., Wu, W., Lee, Y. S., Lee, I., & Bao, X. (2020). The importance of AGO 1 and 4 in post-transcriptional gene regulatory function of tRF5-GluCTC, an respiratory syncytial virus-induced tRNA-derived RNA fragment. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms21228766
Yu, M., Lu, B., Zhang, J., Ding, J., Liu, P., & Lu, Y. (2020). tRNA-derived RNA fragments in cancer: Current status and future perspectives. Journal of Hematology & Oncology, 13, 121. https://doi.org/10.1186/s13045-020-00955-6
Dhahbi, J. M., Spindler, S. R., Atamna, H., Boffelli, D., & Martin, D. I. (2014). Deep sequencing of serum small RNAs identifies patterns of 5’ tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer, 6, 37–47. https://doi.org/10.4137/bic.S20764
Wang, J., Ma, G., Li, M., Han, X., Xu, J., Liang, M., Mao, X., Chen, X., Xia, T., Liu, X., & Wang, S. (2020). Plasma tRNA fragments derived from 5’ ends as novel diagnostic biomarkers for early-stage breast cancer. Mol Ther Nucleic Acids, 21, 954–964. https://doi.org/10.1016/j.omtn.2020.07.026
Nolte-’t Hoen, E. N., Buermans, H. P., Waasdorp, M., Stoorvogel, W., Wauben, M. H., & t Hoen PA,. (2012). Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Research, 40, 9272–9285. https://doi.org/10.1093/nar/gks658
Baglio, S. R., Rooijers, K., Koppers-Lalic, D., Verweij, F. J., Pérez Lanzón, M., Zini, N., Naaijkens, B., Perut, F., Niessen, H. W. M., Baldini, N., & Pegtel, D. M. (2015). Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Research & Therapy, 6, 127. https://doi.org/10.1186/s13287-015-0116-z
Huang, X., Yuan, T., Tschannen, M., Sun, Z., Jacob, H., Du, M., Liang, M., Dittmar, R. L., Liu, Y., Liang, M., Kohli, M., Thibodeau, S. N., Boardman, L., & Wang, L. (2013). Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics, 14, 319. https://doi.org/10.1186/1471-2164-14-319
Dhahbi, J. M., Spindler, S. R., Atamna, H., Boffelli, D., Mote, P., & Martin, D. I. (2013). 5’-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiological Genomics, 45, 990–998. https://doi.org/10.1152/physiolgenomics.00129.2013
Wei, Z., Batagov, A. O., Schinelli, S., Wang, J., Wang, Y., El Fatimy, R., Rabinovsky, R., Balaj, L., Chen, C. C., Hochberg, F., Carter, B., Breakefield, X. O., & Krichevsky, A. M. (2017). Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nature Communications, 8, 1145. https://doi.org/10.1038/s41467-017-01196-x
Lyons, S. M., Gudanis, D., Coyne, S. M., Gdaniec, Z., & Ivanov, P. (2017). Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nature Communications, 8, 1127. https://doi.org/10.1038/s41467-017-01278-w
Gámbaro, F., Li Calzi, M., Fagúndez, P., Costa, B., Greif, G., Mallick, E., Lyons, S., Ivanov, P., Witwer, K., Cayota, A., & Tosar, J. P. (2020). Stable tRNA halves can be sorted into extracellular vesicles and delivered to recipient cells in a concentration-dependent manner. RNA Biology, 17, 1168–1182. https://doi.org/10.1080/15476286.2019.1708548
Duitman, E. H., Orinska, Z., & Bulfone-Paus, S. (2011). Mechanisms of cytokine secretion: A portfolio of distinct pathways allows flexibility in cytokine activity. European Journal of Cell Biology, 90, 476–483. https://doi.org/10.1016/j.ejcb.2011.01.010
Benjamin-Davalos, S., Koroleva, M., Allen, C. L., Ernstoff, M. S., & Shu, S. (2021). Co-isolation of cytokines and exosomes: Implications for immunomodulation studies. Frontiers in Immunology, 12, 638111. https://doi.org/10.3389/fimmu.2021.638111
Pan, L., Huang, X., Liu, Z. X., Ye, Y., Li, R., Zhang, J., Wu, G., Bai, R., Zhuang, L., Wei, L., Li, M., Zheng, Y., Su, J., Deng, J., Deng, S., Zeng, L., Zhang, S., Wu, C., Che, X., … Zheng, J. (2021). Inflammatory cytokine-regulated tRNA-derived fragment tRF-21 suppresses pancreatic ductal adenocarcinoma progression. The Journal of Clinical Investigation. https://doi.org/10.1172/jci148130
Zhang, Y., Liu, Y., Liu, H., & Tang, W. H. (2019). Exosomes: Biogenesis, biologic function and clinical potential. Cell & Bioscience, 9, 19. https://doi.org/10.1186/s13578-019-0282-2
Ratajczak, J., Miekus, K., Kucia, M., Zhang, J., Reca, R., Dvorak, P., & Ratajczak, M. Z. (2006). Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia, 20, 847–856. https://doi.org/10.1038/sj.leu.2404132
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., & Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. https://doi.org/10.1038/ncb1596
Fong, M. Y., Zhou, W., Liu, L., Alontaga, A. Y., Chandra, M., Ashby, J., Chow, A., O’Connor, S. T., Li, S., Chin, A. R., Somlo, G., Palomares, M., Li, Z., Tremblay, J. R., Tsuyada, A., Sun, G., Reid, M. A., Wu, X., Swiderski, P., … Wang, S. E. (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nature Cell Biology, 17, 183–194. https://doi.org/10.1038/ncb3094
Meng, L., Jiang, L., Chen, J., Ren, H., Gao, Z., Wu, F., Wen, Y., & Yang, L. (2021). Transfer RNA-derived fragment tRF-28-QSZ34KRQ590K in plasma exosomes may be a potential biomarker for atopic dermatitis in pediatric patients. Experimental and Therapeutic Medicine, 21, 489. https://doi.org/10.3892/etm.2021.9920
Liu, Y. (2011). Cellular and molecular mechanisms of renal fibrosis. Nature Reviews. Nephrology, 7, 684–696. https://doi.org/10.1038/nrneph.2011.149
Khurana, R., Ranches, G., Schafferer, S., Lukasser, M., Rudnicki, M., Mayer, G., & Hüttenhofer, A. (2017). Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA, 23, 142–152. https://doi.org/10.1261/rna.058834.116
Mami, I., & Pallet, N. (2018). tRNA fragmentation and protein translation dynamics in the course of kidney injury. RNA Biology, 15, 1147–1156. https://doi.org/10.1080/15476286.2015.1107704
Choi, H. R., Steinberg, M. E., & E YC,. (2015). Osteonecrosis of the femoral head: Diagnosis and classification systems. Current Reviews in Musculoskeletal Medicine, 8, 210–220. https://doi.org/10.1007/s12178-015-9278-7
Zhao, J. J., Wu, Z. F., Wang, L., Feng, D. H., & Cheng, L. (2016). MicroRNA-145 mediates steroid-induced necrosis of the femoral head by targeting the OPG/RANK/RANKL signaling pathway. PLoS ONE, 11, e0159805. https://doi.org/10.1371/journal.pone.0159805
Fang, S., He, T., Jiang, J., Li, Y., & Chen, P. (2020). Osteogenic effect of tsRNA-10277-loaded exosome derived from bone mesenchymal stem cells on steroid-induced osteonecrosis of the femoral head. Drug Des Devel Ther, 14, 4579–4591. https://doi.org/10.2147/dddt.S258024
Zhang, Y., Cai, F., Liu, J., Chang, H., Liu, L., Yang, A., & Liu, X. (2018). Transfer RNA-derived fragments as potential exosome tRNA-derived fragment biomarkers for osteoporosis. International Journal of Rheumatic Diseases, 21, 1659–1669. https://doi.org/10.1111/1756-185x.13346
Zhu, L., Li, J., Gong, Y., Wu, Q., Tan, S., Sun, D., Xu, X., Zuo, Y., Zhao, Y., Wei, Y. Q., Wei, X. W., & Peng, Y. (2019). Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Molecular Cancer, 18, 74. https://doi.org/10.1186/s12943-019-1000-8
Lin, C., Zheng, L., Huang, R., Yang, G., Chen, J., & Li, H. (2020). tRFs as potential exosome tRNA-derived fragment biomarkers for gastric carcinoma. Clinical Laboratory. https://doi.org/10.7754/Clin.Lab.2019.190811
Guzman, N., Agarwal, K., Asthagiri, D., Yu, L., Saji, M., Ringel, M. D., & Paulaitis, M. E. (2015). Breast cancer-specific miR signature unique to extracellular vesicles includes “microRNA-like” tRNA fragments. Molecular Cancer Research, 13, 891–901. https://doi.org/10.1158/1541-7786.Mcr-14-0533
Das, S. G., Romagnoli, M., Mineva, N. D., Barillé-Nion, S., Jézéquel, P., Campone, M., & Sonenshein, G. E. (2016). miR-720 is a downstream target of an ADAM8-induced ERK signaling cascade that promotes the migratory and invasive phenotype of triple-negative breast cancer cells. Breast Cancer Research, 18, 40. https://doi.org/10.1186/s13058-016-0699-z
Lu, C., Wang, D., Feng, Y., Feng, L., & Li, Z. (2021). miR-720 regulates insulin secretion by targeting Rab35. BioMed Research International, 2021, 6662612. https://doi.org/10.1155/2021/6662612
Ren, B., Yang, B., Li, P., & Ge, L. (2020). Upregulation of MiR-1274a is correlated with survival outcomes and promotes cell proliferation, migration, and invasion of colon cancer. Oncotargets and Therapy, 13, 6957–6966. https://doi.org/10.2147/ott.S246160
Yoshino, H., Yonezawa, T., Yonemori, M., Miyamoto, K., Sakaguchi, T., Sugita, S., Osako, Y., Tatarano, S., Nakagawa, M., & Enokida, H. (2018). Downregulation of microRNA-1274a induces cell apoptosis through regulation of BMPR1B in clear cell renal cell carcinoma. Oncology Reports, 39, 173–181. https://doi.org/10.3892/or.2017.6098
Ghafour, A. A., Odemis, D. A., Tuncer, S. B., Kurt, B., Saral, M. A., Erciyas, S. K., Erdogan, O. S., Celik, B., Saip, P., & Yazici, H. (2021). High expression level of miR-1260 family in the peripheral blood of patients with ovarian carcinoma. Journal of Ovarian Research, 14, 131. https://doi.org/10.1186/s13048-021-00878-x
Tarazona, R., Delgado, E., Guarnizo, M. C., Roncero, R. G., Morgado, S., Sánchez-Correa, B., Gordillo, J. J., Dejulián, J., & Casado, J. G. (2011). Human prostasomes express CD48 and interfere with NK cell function. Immunobiology, 216, 41–46. https://doi.org/10.1016/j.imbio.2010.03.002
Vojtech, L., Woo, S., Hughes, S., Levy, C., Ballweber, L., Sauteraud, R. P., Strobl, J., Westerberg, K., Gottardo, R., Tewari, M., & Hladik, F. (2014). Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Research, 42, 7290–7304. https://doi.org/10.1093/nar/gku347
Goodfellow, S. J., & White, R. J. (2007). Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle, 6, 2323–2326. https://doi.org/10.4161/cc.6.19.4767
Liapi, E., van Bilsen, M., Verjans, R., & Schroen, B. (2020). tRNAs and tRNA fragments as modulators of cardiac and skeletal muscle function. Biochimica et Biophysica Acta, Molecular Cell Research, 1867, 118465. https://doi.org/10.1016/j.bbamcr.2019.03.012
Zhu, X. L., Li, T., Cao, Y., Yao, Q. P., Liu, X., Li, Y., Guan, Y. Y., Deng, J. J., Jiang, R., & Jiang, J. (2021). tRNA-derived fragments tRF(GlnCTG) induced by arterial injury promote vascular smooth muscle cell proliferation. Mol Ther Nucleic Acids, 23, 603–613. https://doi.org/10.1016/j.omtn.2020.12.010
Fu, S., Zhang, Y., Li, Y., Luo, L., Zhao, Y., & Yao, Y. (2020). Extracellular vesicles in cardiovascular diseases. Cell Death Discov, 6, 68. https://doi.org/10.1038/s41420-020-00305-y
Weng, Q., Wang, Y., Xie, Y., Yu, X., Zhang, S., Ge, J., Li, Z., Ye, G., & Guo, J. (2022). Extracellular vesicles-associated tRNA-derived fragments (tRFs): Biogenesis, biological functions, and their role as potential biomarkers in human diseases. Journal of Molecular Medicine (Berlin, Germany), 100, 679–695. https://doi.org/10.1007/s00109-022-02189-0
Xie, Y., Yao, L., Yu, X., Ruan, Y., Li, Z., & Guo, J. (2020). Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduction and Targeted Therapy, 5, 109. https://doi.org/10.1038/s41392-020-00217-4
Acknowledgements
TV acknowledges Central University of Kerala for support. PSS thank Faculty seed research grant of the National Institute of Technology, Calicut and Department of Biotechnology (BT/PR16307/MED/30/1729/2016) Govt of India for the support. ST thank Ministry of Education, Govt of India and the National Institute of Technology, Calicut for the fellowship. We apologize to many scientists whose research studies were not included in this review due to page/word limit.
Author information
Authors and Affiliations
Contributions
TV conceptualized and drafted the manuscript. ST contributed to figure preparation and editing of the manuscript. PSS drafted, edited, and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed Consent
Not applicable.
Research Involving Human and Animal Rights
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suresh, P.S., Thankachan, S. & Venkatesh, T. Landscape of Clinically Relevant Exosomal tRNA-Derived Non-coding RNAs. Mol Biotechnol 65, 300–310 (2023). https://doi.org/10.1007/s12033-022-00546-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00546-5