Abstract
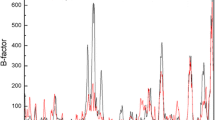
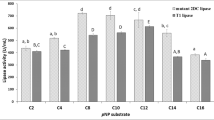
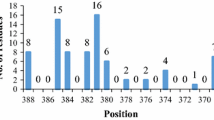
A novel lipase has been recently isolated from a local Pseudomonas sp. (GQ243724). In the present study, we have tried to increase the organic solvent stability of this lipase using site-directed mutagenesis. Eight variants N219L, N219I, N219P, N219A, N219R, N219D, S251L, and S251K were designed to change the surface hydrophobicity of this enzyme with respect to the wild-type. Among these variants, the stability of N219L and N219I significantly increased in the presence of all tested organic solvents, whereas two mutants (N219R and N219D) significantly exhibited decreased stabilities in all the organic solvent studied, suggesting that improvement of hydrophobic patches on the enzyme surface enhances the stability in organic media. Furthermore, replacing Ser251 with hydrophobic residues on the enzyme surface dramatically diminished its stability in the tested condition. In spite of the distance of the mutated sites from the active site, the values of k cat and K m were affected. Finally, structural analysis of the wild-type and mutated variants was carried out in the presence and absence of some organic solvents using circular dichroism and fluorescence spectroscopy.




Similar content being viewed by others
References
Arnold, F. H. (1990). Enzyme engineering for non-aqueous media. Trends in Biotechnology, 8, 244–249.
Doukyu, N., & Ogino, H. (2010). Organic solvent-tolerant enzymes. Biochemical Engineering Journal, 48, 270–282.
Ogino, H., Uchiho, T., Doukyu, N., Yasuda, M., Ishimi, k, & Ishikawa, H. (2007). Effect of exchange of amino acid residues of the surface region of the PST-01 protease on its organic solvent-stability. Biochemical and Biophysical Research Communications, 358, 1028–1033.
Arnold, F. H. (1988). Protein design for non-aqueous solvents. Protein Engineering, 2, 21–25.
Gupta, A., Ray, S., Kapoor, S., & Khare, S. K. (2008). Solvent-stable Pseudomonas aeruginosa PseA protease gene: Identification, molecular characterization, phylogenetic and bioinformatic analysis to study reasons for solvent stability. Journal of Molecular Microbiology and Biotechnology, 15, 234–243.
Gaur, R., Grover, T., Sharma, R., Kapour, S., & Khare, S. (2010). Purification and characterization of a solvent stable aminopeptidase from Pseudomonas aeruginosa: Cloning and analysis of amino peptidase gene conferring solvent stability. Process Biochemistry, 45, 757–764.
Chakravorty, D., Parameswaran, S., Dubey, V. K., & Patra, S. (2012). Unraveling the rationale behind organic solvent stability of lipases. Applied Biochemistry and Biotechnology, 167, 439–461.
Tripathi, M. K., Roy, U., Jinwal, U. K., Jain, S. K., & Roy, P. K. (2004). Cloning, sequencing and structural features of a novel Streptococcus lipase. Enzyme and Microbial Technology, 34, 437–445.
Yu, M., Qin, S., & Tan, T. (2007). Purification and characterization of the extracellular lipase Lip2 from Yarrowia lipolytica. Process Biochemistry, 42, 384–391.
Gromiha, M. M., Oobatake, M., Kono, H., Uedaira, H., & Sarai, A. (2002). Importance of mutant position in Ramachandran plot for predicting protein stability of surface mutations. Biopolymers, 64, 210–220.
Bava, K. A., Gromiha, M. M., Uedaira, H., Kitajima, K., & Sarai, A. (2004). Protherm, version 4.0. Thermodynamic database for proteins and mutants. Nucleic Acids Research, 32, D120–D121.
Akbari, N., Daneshjoo, S., Akbari, J., & Khajeh, K. (2011). Isolation, characterization and catalytic properties of a novel lipase which is activated in ionic liquids and organic solvents. Applied Biochemistry and Biotechnology, 165, 785–794.
Akbari, N., Khajeh, K., Rezaie, S., Mirdamadi, S., & Shavandi, M. (2009). High level expression of lipase in Escherichia coli and recovery of active recombinant enzyme through in vitro refolding. Protein Expression and Purification, 70, 75–80.
Nardini, M., Lang, D. A., Liebeton, K., Jaeger, K. E., & Dijkstra, B. W. (2000). Crystal structure of Pseudomonas aeruginosa lipase in the open conformation. Journal of Biological Chemistry, 275, 31219–31225.
Martinez, P., Van Dam, M. E., Robinson, A. C., Chen, K., & Arnold, F. H. (1992). Stabilization of subtilisin E in organic solvents by site-directed mutagenesis. Biotechnology and Bioengineering, 39, 141–147.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J. Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
Dantas, G., Corrent, C., Reichow, S. L., Havranek, J. J., Eletr, Z. M., Isern, N. G., et al. (2007). High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. Journal of Molecular Biology, 366, 1209–1221.
Kawata, T., & Ogino, H. (2010). Amino acid residues involved in organic solvent-stability of the LST-03 lipase. Biochemical and Biophysical Research Communications, 400, 348–388.
Yang, S., Zhou, L., Tang, H., Pan, J., Wu, X., Huang, H., et al. (2002). Rational design of a more stable penicillin G acylase against organic cosolvent. Journal of Molecular Catalysis B: Enzymatic, 18, 258–290.
Fisher, C. L., & Pei, G. K. (1997). Modification of a PCR-based site-directed mutagenesis method. BioTechniques, 23, 570–574.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Winkler, U. K., & Stuckman, M. (1979). Glycogen, hyaluronate and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. Journal of Bacteriology, 3, 663–670.
Gupta, N., Rathi, P., & Gupta, R. (2002). Simplified para-nitrophenyl palmitate assay for lipases and esterases. Analytical Biochemistry, 311, 98–99.
Karadzic, I., Masui, A., Zivkovic, L. I., & Fujiwara, N. (2006). Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metal working fluid. Journal of Bioscience and Bioengineering, 102, 82–89.
Eftink, M. R., & Ghiron, C. A. (1977). Exposure of tryptophanyl residues and protein dynamics. Biochemistry, 16, 5546–5551.
Kelly, S. M., Jess, T. J., & Price, N. (2005). How to study proteins by circular dichroism. Biochimica et Biophysica Acta, 1751, 119–139.
Ogino, H., Nakagawa, S., Shinya, T., Muto, T., Fujimura, M., Yasudo, K., et al. (2000). Purification and characterization of organic solvent-tolerance lipase from organic solvent-tolerant Pseudomonas aeruginosa LST-03. Journal of Bioscience and Bioengineering, 89, 451–457.
Sugihara, A., Ueshima, M., Shimada, Y., Tsunasawa, S., & Tominaga, Y. (1992). Purification and characterization of a novel thermostable lipase from Pseudomonas cepacia. Journal of Biochemistry, 112, 598–603.
Sharma, A. K., Ti, W., Ari, R. P., & Hoondal, G. (2001). Properties of a thermostable and solvent stable extracellular lipase from a Pseudomonas sp.AG.8. Journal of Basic Microbiology, 41, 363–366.
Gang, W., Gung, C., & Ming, W. (2011). Biochemical properties and potential applications of an organic solvent-tolerant lipase isolated from Bacillus cereus BF-3. African Journal of Biotechnology, 10, 13174–13179.
Lakowicz, J. R. (1992). Topics in fluorescence spectroscopy-biological applications (p. 3). New York: Plenum Press.
Figueiredo, K. C. S., Ferraz, H. C., Borges, C. P., & Alves, T. L. M. (2009). Structural stability of myoglobin in organic media. Protein Journal, 28, 224–232.
Yadavalli, P., & Rao, N. M. (2013). Engineering the loops in a lipase for stability in DMSO. Protein Engineering Design and Selection, 26, 317–324.
Ogino, H., Gemba, Y., Yutori, Y., Doukyu, N., Ishimi, K., & Ishikawa, H. (2007). Stabilities and conformational transitions of various proteases in the presence of an organic solvent. Biotechnology Progress, 23, 155–161.
Chandrayan, S. K., Dhaunta, N., & Guptasarma, p. (2008). Expression, purification, refolding and characterization of a putative lysophospholipase from Pyrococcus furiosus: Retention of structure and lipase/esterase activity in the presence of water-miscible organic solvents at high temperatures. Protein Expression and Purification, 59, 327–333.
Cowan, D. A. (1977). Thermophilic proteins: Stability and function in aqueous and organic solvents. Comparative Biochemistry and Physiology, 118A, 429–438.
Hao, J., & Berry, A. (2004). A thermostable variant of fructose bisphosphate aldolase constructed by directed evolution also show increased stability in organic solvents. Protein Engineering, Design and Selection, 17, 689–697.
Vasquez-Figueroa, E., Yah, V., Broering, J. M., Chaparro-Riggers, J. F., & Bommarius, A. S. (2008). Thermostable variants constructed via the structure-guided consensus method also show increased stability in salts solutions and homogeneous aqueous-organic media. Protein Engineering, Design and Selection, 21, 673–680.
Acknowledgments
The authors would like to express their gratitude to the Research Council of the University of Tehran and Tarbiat Modares University for their financial support.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Monsef Shokri, M., Ahmadian, S., Akbari, N. et al. Hydrophobic Substitution of Surface Residues Affects Lipase Stability in Organic Solvents. Mol Biotechnol 56, 360–368 (2014). https://doi.org/10.1007/s12033-013-9716-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-013-9716-y