Abstract
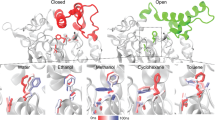
Organic solvent-stable lipases have pronounced impact on industrial economy as they are involved in synthesis by esterification, interesterification, and transesterification. However, very few of such natural lipases have been isolated till date. A study of the recent past provided few pillars to rely on for this work. The three-dimensional structure, inclusive of the surface and active site, of 29 organic solvent-stable lipases was analyzed by subfamily classification and protein solvent molecular docking based on fast Fourier transform correlation approach. The observations revealed that organic solvent stability of lipases is their intrinsic property and unique with respect to each lipase. In this paper, factors like surface distribution of charged, hydrophobic, and neutral residues, interaction of solvents with catalytically immutable residues, and residues interacting with essential water molecules required for lipase activity, synergistically and by mutualism contribute to render a stable lipase organic solvent. The propensity of surface charge in relation to stability in organic solvents by establishing repulsive forces to exclude solvent molecules from interacting with the surface and prohibiting the same from gaining entry to the protein core, thus stabilizing the active conformation, is a new finding. It was also interesting to note that lipases having equivalent surface-exposed positive and negative residues were stable in a wide range of organic solvents, irrespective of their LogP values.





Similar content being viewed by others
Abbreviations
- ASA:
-
Accessible surface area
- SE:
-
Surface exposed
References
Jaeger, K. E., & Reetz, M. T. (1998). Microbial lipases form versatile tools for biotechnology. Trends in Biotechnology, 16(9), 396–403.
Ogino H. (2008) Protein adaptation in extremophiles. In: Siddiqui K.S., Thomas T (eds). Nova Science, New York, pp. 193–236.
Hun, C. J., Rahman, R. N. Z. A., Salleh, A. B., & Basri, M. A. (2003). Newly isolated organic solvent tolerant Bacillus sphaericus 205y producing organic solvent-stable lipase. Biochemical Engineering Journal, 15(2), 147–151.
Ogino, H., & Ishikawa, H. (2001). Enzymes which are stable in the presence of organic solvents. Journal of Bioscience and Bioengineering, 91, 109–116.
Secundo, F., & Carrea, G. (2002). Lipase activity and conformation in neat organic solvents. Journal of Molecular Catalysis B: Enzymatic, 19, 93–102.
Hasan, F., Shah, A. A., & Hameed, A. (2006). Industrial applications of microbial lipases. Enzyme and Microbial Technology, 39(2), 235–251.
Cardenas, F., de Castro, M. S., Sanchez-Montero, J. M., Sinisterra, J. V., Valmaseda, M., Elson, S. W., et al. (2001). Novel microbial lipases: catalytic activity in reactions in organic media. Enzyme and Microbial Technology, 28, 145–154.
Fang, Y., Lu, Z., Lv, F., Bie, X., Liu, S., Ding, Z., et al. (2006). Newly isolated organic solvent tolerant Staphylococcus saprophyticus M36 produced organic solvent-stable lipase. Current Microbiology, 53(6), 510–515.
Timasheff, S. N. (1993). The control of protein stability and association by weak interactions with water: how do solvents affect these processes? Annual Review of Biophysics and Biomolecular Structure, 22, 67–97.
Schulze, B., & Klibanov, A. M. (1991). Inactivation and stabilization of stabilizing in neat organic solvents. Biotechnology and Bioengineering, 38(9), 1001–1006.
Martinez, P., & Arnold, F. H. (1991). Surface charge substitutions increase the stability of alpha- lytic protease in organic solvents. Journal of the American Chemical Society, 113(16), 6336–6337.
Kumar, M., Thakur, V., & Raghava, G. P. S. (2010). COPid: composition based protein identification. In Silico Biology, 8, 11.
Kelil, A., Wang, S., Brzezinski, R., & Fleury, A. (2007). CLUSS: clustering of protein sequences based on a new similarity measure. BMC Bioinformatics, 8, 1–19.
Roy, A., Kucukural, A., & Yang, Z. (2009). I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, 5, 725–738.
Ahmad, S., Gromiha, M. M., Fawareh, H., & Sarai, A. (2004). ASAView: solvent accessibility graphics for proteins. BMC Bioinformatics, 5, 51.
Pavelka, A., Chovancova, E., & Damborsky, J. (2009). HotSpot wizard: a web server for identification of hot spots in protein engineering. Nucl Acids Res, 37, W376–W383.
Brenke, R., Kozakov, D., Chuang, G.-Y., Beglov, D., Mattos, C., & Vajda, S. (2009). Fragment-based identification of druggable "hot spots" of proteins using Fourier domain correlation. Bioinformatics, 25(5), 621–627.
Díaz-García, M. E., & Valencia-González, M. J. (1995). Enzyme catalysis in organic solvents: a promising field for optical biosensing. Talanta, 42(11), 1763–1773.
Kuntz, I. D., & Kauzmann, W. (1974). Hydration of proteins and polypeptides. Advances in Protein Chemistry, 28, 239–345.
Valivety, R. H., Halling, P. J., & Macrae, A. R. (1992). Reaction rate with lipase catalyst shows similar dependence on water activity in different organic solvents. Biochimica et Biophysica Acta, 1118, 218–222.
Arakawa, T., Kita, Y., & Timasheff, S. N. (2007). Protein precipitation and denaturation by dimethyl sulfoxide. Biophysical Chemistry, 131, 62–70.
Nagao, T., Shimada, Y., Sugihara, A., & Tominaga, Y. (2002). Increase in stability of Fusarium heterosporum lipase. Journal of Molecular Catalysis B: Enzymatic, 17, 125–132.
Sugihara, A., Ueshima, M., Shimada, Y., Tsunasawa, S., & Tominaga, Y. (1992). Purification and characterization of a novel thermostable lipase from Pseudomonas cepacia. Journal of Biochemistry, 112(5), 598–603.
Choo, D. W., Kurihara, T., Suzuki, T., Soda, K., & Esaki, N. (1998). A cold-adapted lipase of an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: gene cloning and enzyme purification and characterization. Applied and Environmental Microbiology, 64(2), 486–491.
Yan, J., Yang, J., Xu, L., & Yan, Y. (2007). Gene cloning, overexpression and characterization of a novel organic solvent tolerant and thermostable lipase from Galactomyces geotrichum Y05. Journal of Molecular Catalysis B: Enzymatic, 49, 28–35.
Yu, M., Qin, S., & Tan, T. (2007). Purification and characterization of the extracellular lipase Lip2 from Yarrowia lipolytica. Protein Biochemistry, 42, 384–391.
Lescic, I., Vukelic, B., Elenkov, M. M., Saenger, W., & Abramic, M. (2001). Substrate specificity and effects of water-miscible solvents on the activity and stability of extracellular lipase from Streptomyces rimosus. Enzyme and Microbial Technology, 29, 548–553.
Soliman, N. A., Knoll, M., Abdel-Fattah, Y., Schmid, R. D., & Lange, S. (2007). Molecular cloning and characterization of thermostable esterase and lipase from Geobacillus thermoleovorans YN isolated from desert soil in Egypt. Protein Biochemistry, 42(7), 1090–1100.
Wasylewski, Z., & Kozik, A. (1979). Protein–non-ionic detergent interaction. Interaction of bovine serum albumin with alkyl glucosides studied by equilibrium dialysis and infrared spectroscopy. European Journal of Biochemistry, 95, 121–126.
Lad, M. D., Ledger, V. M., Briggs, B., Green, R. J., & Frazier, R. A. (2003). Analysis of the SDS–lysozyme binding isotherm. Langmuir, 19(12), 5098–5103.
Salameh, M. A., & Wiegel, J. (2010). Effects of detergents on activity, thermostability and aggregation of two alkalithermophilic lipases from Thermosyntropha lipolytica. Open Biochemical Journal, 4, 22–28.
Anfinsen, C.B. (1970). Advances in protein chemistry. In: Richards, F.M. (ed). Academic Press, London, vol. 24, pp. 41.
Gekko, K., & Timasheff, S. N. (1981). Mechanism of protein stabilization by glycerol: preferential hydration in glycerol–water mixtures. Biochemistry, 20, 4667–4676.
Gekko, K., Ohmae, E., Kameyama, K., & Takagi, T. (1998). Acetonitrile-protein interactions: amino acid solubility and preferential solvation. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology, 1387(1–2), 195–205.
Gromiha, M. M., Santhosh, C., & Ahmad, S. (2004). Structural analysis of cation-pi interactions in DNA binding proteins. International Journal of Biological Macromolecules, 34, 203–211.
Irun, M. P., Maldonado, S., & Sancho, J. (2001). Stabilization of apoflavodoxin by replacing hydrogen-bonded charged Asp or Glu residues by the neutral isosteric Asn or Gln. Protein Engineering, 14(3), 173–181.
Madan, B., & Mishra, P. (2009). Overexpression, purification and characterization of organic solvent stable lipase from Bacillus licheniformis RSP-09. Journal of Molecular Microbiology and Biotechnology, 17, 118–123.
Quyen, D. T., Nguyen, T. T., Le, T. T., Kim, H. K., Oh, T. K., & Lee, J. K. (2004). A novel lipase/chaperone pair from Ralstonia sp. M1: analysis of the folding interaction and evidence for gene loss in R. solanacearum. Molecular Genetics and Genomics, 272(5), 538–549.
Gulati, R., Saxena, R. K., Gupta, R., Yadav, R. P., & Davidson, W. S. (1999). Parametric optimization of Aspergillus terreus lipase production and its potential in ester synthesis. Protein Biochemistry, 35(5), 459–464.
Zhang, M., Stauffacher, C. V., Linand, D., & Etten, R. L. (1998). Crystal structure of a human low molecular weight phosphotyrosyl phosphatase implications for substrate specificity. Journal of Biological Chemistry, 273, 21714–21720.
Zhao, L. L., Xua, J. H., Zhaoa, J., Pana, J., & Wangb, Z. L. (2008). Biochemical properties and potential applications of an organic solvent-tolerant lipase isolated from Serratia marcescens ECU1010. Protein Biochemistry, 43(6), 626–633.
Tripathi, M. K., Roy, U., Jinwal, U. K., Jain, S. K., & Roy, P. K. (2004). Cloning, sequencing and structural features of a novel Streptococcus lipase. Enzyme and Microbial Technology, 34, 437–445.
Dellamora-Ortiz, G. M., Martins, R. C., Rocha, W. L., & Dias, A. P. (1997). Activity and stability of a Rhizomucor miehei lipase in hydrophobic media. Biotechnology and Applied Biochemistry, Pt 1, 31–37.
Sulong, M. R., Abdul Rahman, R. N., Salleh, A. B., & Basri, M. A. (2006). Novel organic solvent tolerant lipase from Bacillus sphaericus 205y: extracellular expression of a novel OST-lipase gene. Protein Expression and Purification, 49(2), 190–195.
Ogino, H., Miyamoto, M., Yasuda, M., Ishimi, K., & Ishikawa, H. (1999). Growth of organic solvent-tolerant Pseudomonas aeruginosa LST-03 in the presence of various organic solvents and production of lipolytic enzyme in the presence of cyclohexane. Biochemical Engineering Journal, 4(1), 1–6.
Rahman, R. N., Baharum, S. N., Salleh, A. B., & Basri, M. (2005). S5 lipase: an organic solvent tolerant enzyme. Journal of Microbiology, 44(6), 583–590.
Horchani, H., Mosbah, H., Salem, N. B., Gargouri, Y., & Sayari, A. (2008). Biochemical and molecular characterisation of a thermoactive, alkaline and detergent-stable lipase from a newly isolated Staphylococcus aureus. Journal of Molecular Catalysis B: Enzymatic, 56(4), 237–245.
Eltaweel, M. A., Rahman, R. A., Salleh, A. B., & Basri, M. (2005). An organic solvent stable lipase from Bacillus sp. strain 42. Annals of Microbiology, 55(3), 187–192.
Hiol, A., Jonzo, M. D., Rugani, N., Druet, D., Sarda, L., & Comeau, L. C. (2000). Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzyme and Microbial Technology, 26, 421–430.
Tang, Y., Lu, Y., Lu, F., Bie, X., Guo, Y., & Lu, Z. (2009). Cloning and expression of organic solvent tolerant lipase gene from Staphylococcus saprophyticus M36. Sheng Wu Gong Cheng Xue Bao, 25(12), 1989–1995.
Sinchaikul, S., Tyndall, J. D. A., Fothergill-Gilmore, L. A., Taylor, P., Phutrakul, S., Chen, S. T., et al. (2002). Expression, purification, crystallization and preliminary crystallographic analysis of a thermostable lipase from Bacillus stearothermophilus P1. Acta Crystallographica, D58, 182–185.
Schmidt-Dannert, C., Rua, M. L., Atomi, H., & Schmid, R. D. (1996). Thermoalkalophilic lipase of Bacillus thermocatenulatus molecular cloning, nucleotide sequence, purification and some properties. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism, 1301, 105–114.
Li, H., & Zhang, X. (2005). Characterization of thermostable lipase from thermophilic Geobacillus sp. TW1. Protein Expression and Purification, 42(1), 153–159.
Liu, S., Fang, Y., Xu, W., Lu, M., Wang, S., & Chen, L. (2009). Screening and identification of a novel organic solvent-stable lipase producer. Annals of Microbiology, 59(3), 539–543.
Iizumi, T., Nakamura, K., & Fukase, T. (1990). Purification and characterization of a thermostable lipase from newly isolated Pseudomonas sp. KWI-56. Agricultural Biological Chemistry, 54(5), 1253–1258.
Royter, M., Schmidt, M., Elend, C., Hobenreich, H., Schafer, T., Bornscheuer, U. T., et al. (2009). Thermostable lipases from the extreme thermophilic anaerobic bacteria Thermoanaerobacter thermohydrosulfuricus SOL1 and Caldanaerobacter subterraneus subsp. tengcongensis. Extremophiles, 13(5), 769–783.
Shu, Z. Y., Yang, J. K., & Yan, Y. J. (2007). Purification and characterization of a lipase from Aspergillus niger F044. Chinese Journal of Biotechnology, 23, 96–101.
Han, S. J., Back, J. H., Yoon, M. Y., Shin, P. K., Cheong, C. S., Sung, M. H., et al. (2003). Expression and characterization of a novel enantioselective lipase from Acinetobacter species SY-01. Biochimie, 85, 501–510.
Acknowledgments
This work was supported by the Department of Information Technology, Government of India.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Material Table 1
Details of result as mentioned in text are provided in supplementary Table 1 highlights the ten annotated subfamilies of the twenty nine organic solvent stable lipases laying especial emphasis on the surface amino acid residue composition. (DOC 111 kb)
Rights and permissions
About this article
Cite this article
Chakravorty, D., Parameswaran, S., Dubey, V.K. et al. Unraveling the Rationale Behind Organic Solvent Stability of Lipases. Appl Biochem Biotechnol 167, 439–461 (2012). https://doi.org/10.1007/s12010-012-9669-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-012-9669-9