Abstract
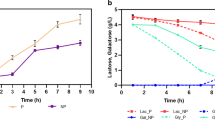
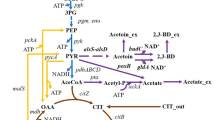
Over-expression of recombinant proteins in Escherichia coli triggers a metabolic stress response which causes a sharp decline in both growth and product formation rates post induction. We identified a key down-regulated substrate utilization gene, glycerol kinase (glpK), whose up-regulation could help alleviate this stress response. In a proof of principal study conducted in shake flask cultures, the glpK gene under the “ara” promoter in a pPROLar.A122 vector was co-transformed along with the recombinant interferon-β (rhIFN-β) gene in a pET22b vector into E. coli BL-21(DE3) cells. Co-expression of glpK improved the expression levels of rhIFN-β in glycerol containing medium, while no such gain was observed in medium without glycerol. This study was extended to high cell density fed-batch cultures where exponential feeding of complex substrates was done to increase biomass and hence product titers. For this we first constructed a modified E. coli strain BL-21(glpK +) where the glpK gene was inserted downstream of the ibpA promoter in the host chromosome. There was a significant improvement in growth as well as expression levels of rhIFN-β in this modified strain when the feed medium contained high glycerol. A final product concentration of 4.8 g/l of rhIFN-β was obtained with the modified strain which was 35 % higher than the control.







Similar content being viewed by others
References
Yee, L., & Blanch, H. W. (1992). Recombinant protein expression in high cell density fed-batch cultures of Escherichia coli. Nature Biotechnology, 10, 1550–1556.
Cheng, L. -C., Hor, L. -I., Wu, J. -Y., & Chen, T. -L. (2003). Effect of specific growth rate on the production of a recombinant nuclease by Escherichia coli. Biochemical Engineering Journal, 14, 101–107.
Choi, J. H., Keum, K. C., & Lee, S. Y. (2006). Production of recombinant proteins by high cell density culture of Escherichia coli. Chemical Engineering Science, 61, 876–885.
Matsui, T., Sato, H., Yamamuro, H., Misawa, S., Shinzato, N., Matsuda, H., et al. (2008). High cell density cultivation of recombinant Escherichia coli for hirudin variant 1 production. Journal of Biotechnology, 134, 88–92.
Srivastava, P., Bhattacharaya, P., Pandey, G., & Mukherjee, K. J. (2005). Overexpression and purification of recombinant human interferon alpha2b in Escherichia coli. Protein Expression Purification, 41, 313–322.
Seo, J. H., Kang, D. G., & Cha, H. J. (2003). Comparison of cellular stress levels and green-fluorescent-protein expression in several Escherichia coli strains. Biotechnology and Applied Biochemistry, 37, 103–107.
Hoffmann, F., & Rinas, U. (2004). Stress induced by recombinant protein production in Escherichia coli. Advances in Biochemical Engineering/Biotechnology, 89, 73–92.
Choi, J. H., Lee, S. J., & Lee, S. Y. (2003). Enhanced production of insulin-like growth factor I fusion protein in Escherichia coli by coexpression of the down-regulated genes identified by transcriptome profiling. Applied and Environment Microbiology, 69, 4737–4742.
Durrschmid, K., Reischer, H., Schmidt-Heck, W., Hrebicek, T., Guthke, R., Rizzi, A., et al. (2008). Monitoring of transcriptome and proteome profiles to investigate the cellular response of E. coli towards recombinant protein expression under defined chemostat conditions. Journal of Biotechnology, 135, 34–44.
Ow, D. S. -W., Nissom, P. M., Philp, R., Oh, S. K. -W., & Yap, M. G. -S. (2006). Global transcriptional analysis of metabolic burden due to plasmid maintenance in Escherichia coli DH5α during batch fermentation. Enzyme and Microbial Technology, 39, 391–398.
Lee, K. H., Park, J. H., Kim, T. Y., Kim, H. U., & Lee, S. Y. (2007). Systems metabolic engineering of Escherichia coli for l-threonine production. Molecular Systems Biology, 3, 149.
March, J. C., Eiteman, M. A., & Altman, E. (2002). Expression of an anaplerotic enzyme, pyruvate carboxylase, improves recombinant protein production in Escherichia coli. Applied and Environment Microbiology, 68, 5620–5624.
Weikert, C., Canonaco, F., Sauer, U., & Bailey, J. E. (2000). Co-overexpression of RspAB improves recombinant protein production in Escherichia coli. Metabolic Engineering, 2, 293–299.
Li, Y. H., Liu, Y., Wang, S. C., Tong, Z. Y., & Xu, Q. S. (2003). Co-expressions of phosphoenolpyruvate synthetase A (ppsA) and transketolase A (tktA) genes of Escherichia coli. Sheng Wu Gong Cheng Xue Bao, 19, 301–306.
Chou, C. (2007). Engineering cell physiology to enhance recombinant protein production in Escherichia coli. Applied Microbiology and Biotechnology, 76, 521–532.
Han, M. -J., Park, S. J., Park, T. J., & Lee, S. Y. (2004). Roles and applications of small heat shock proteins in the production of recombinant proteins in Escherichia coli. Biotechnology and Bioengineering, 88, 426–436.
Harcum, S. W., & Haddadin, F. T. (2006). Global transcriptome response of recombinant Escherichia coli to heat-shock and dual heat-shock recombinant protein induction. Journal of Industrial Microbiology and Biotechnology, 33, 801–814.
Flores, S., Flores, N., de Anda, R., Gonzalez, A., Escalante, A., Sigala, J. C., et al. (2005). Nutrient-scavenging stress response in an Escherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system, as explored by gene expression profile analysis. Journal of Molecular Microbiology and Biotechnology, 10, 51–63.
Singh, A. B., Sharma, A. K., & Mukherjee, K. J. (2012). Analyzing the metabolic stress response of recombinant Escherichia coli cultures expressing human interferon-beta in high cell density fed batch cultures using time course transcriptomic data. Molecular BioSystems, 8, 615–628.
Link, A. J., Phillips, D., & Church, G. M. (1997). Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. Journal of Bacteriology, 179, 6228–6237.
Maniatis, T., Fritsch, E. F., Sambrook, J. (1982) Molecular cloning: A laboratory manual In T. Maniatis, E. F. Fritsch & J. Sambrook (Eds.) Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory.
Beuken, E., Vink, C., & Bruggeman, C. A. (1998). One-step procedure for screening recombinant plasmids by size. BioTechniques, 24, 748–750.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Lee, J., Lee, S. Y., Park, S., & Middelberg, A. P. (1999). Control of fed-batch fermentations. Biotechnology Advances, 17, 29–48.
Lopez, M. F., Berggren, K., Chernokalskaya, E., Lazarev, A., Robinson, M., & Patton, W. F. (2000). A comparison of silver stain and SYPRO ruby protein gel stain with respect to protein detection in two-dimensional gels and identification by peptide mass profiling. Electrophoresis, 21, 3673–3683.
Nolan, T., Hands, R. E., & Bustin, S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nature Protocols, 1, 1559–1582.
Beijer, L., Nilsson, R. P., Holmberg, C., & Rutberg, L. (1993). The glpP and glpF genes of the glycerol regulon in Bacillus subtilis. Journal of General Microbiology, 139, 349–359.
Zwaig, N., Kistler, W. S., & Lin, E. C. (1970). Glycerol kinase, the pacemaker for the dissimilation of glycerol in Escherichia coli. Journal of Bacteriology, 102, 753–759.
Charrier, V., Buckley, E., Parsonage, D., Galinier, A., Darbon, E., Jaquinod, M., et al. (1997). Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. Journal of Biological Chemistry, 272, 14166–14174.
Gill, R. T., Valdes, J. J., & Bentley, W. E. (2000). A comparative study of global stress gene regulation in response to overexpression of recombinant proteins in Escherichia coli. Metabolic Engineering, 2, 178–189.
Bentley, W. E., Mirjalili, N., Andersen, D. C., Davis, R. H., & Kompala, D. S. (1990). Plasmid-encoded protein: The principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnology and Bioengineering, 35, 668–681.
Glick, B. R. (1995). Metabolic load and heterologous gene expression. Biotechnology Advances, 13, 247–261.
Neubauer, P., Lin, H. Y., & Mathiszik, B. (2003). Metabolic load of recombinant protein production: Inhibition of cellular capacities for glucose uptake and respiration after induction of a heterologous gene in Escherichia coli. Biotechnology and Bioengineering, 83, 53–64.
Wang, Z., Xiang, L., Shao, J., Wegrzyn, A., & Wegrzyn, G. (2006). Effects of the presence of ColE1 plasmid DNA in Escherichia coli on the host cell metabolism. Microbial Cell Factories, 5, 34.
Boyd, D., Weiss, D. S., Chen, J. C., & Beckwith, J. (2000). Towards single-copy gene expression systems making gene cloning physiologically relevant: Lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. Journal of Bacteriology, 182, 842–847.
Martinez-Morales, F., Borges, A. C., Martinez, A., Shanmugam, K. T., & Ingram, L. O. (1999). Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons used during construction. Journal of Bacteriology, 181, 7143–7148.
Dragosits, M., Nicklas, D., & Tagkopoulos, I. (2012). A synthetic biology approach to self-regulatory recombinant protein production in Escherichia coli. Journal of Biological Engineering, 6, 2.
Gosset, G., Anda, R., Cruz, N., Martínez, A., Quintero, R., & Bolivar, F. (1993). Recombinant protein production in cultures of an Escherichia coli trp-strain. Applied Microbiology and Biotechnology, 39, 541–546.
Aristidou, A. A., San, K. Y., & Bennett, G. N. (1995). Metabolic engineering of Escherichia coli to enhance recombinant protein production through acetate reduction. Biotechnology Progress, 11, 475–478.
Vemuri, G. N., Eiteman, M. A., & Altman, E. (2006). Increased recombinant protein production in Escherichia coli strains with overexpressed water-forming NADH oxidase and a deleted ArcA regulatory protein. Biotechnology and Bioengineering, 94, 538–542.
Picon, A., de Mattos, M. J., & Postma, P. W. (2008). Protein production by Escherichia coli wild-type and Delta ptsG mutant strains with IPTG induction at the onset. Journal of Industrial Microbiology and Biotechnology, 35, 213–218.
Acknowledgments
Financial support by Department of Biotechnology, Government of India is gratefully acknowledged. We would also like to kindly acknowledge Prof Hermann Bujard, (ZMBH) Universitaet Heidelberg for the plasmid pPROLar.A122 and Dr. Sara L. Vassallo, Harvard Medical School, Boston for the plasmid pKOV.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, A.B., Mukherjee, K.J. Supplementation of Substrate Uptake Gene Enhances the Expression of rhIFN-β in High Cell Density Fed-Batch Cultures of Escherichia coli . Mol Biotechnol 54, 692–702 (2013). https://doi.org/10.1007/s12033-012-9611-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-012-9611-y