Abstract
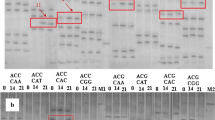
Plant growth and productivity are affected by various abiotic stresses such as heat, drought, cold, salinity, etc. The mechanism of salt tolerance is one of the most important subjects in plant science as salt stress decreases worldwide agricultural production. In our present study we used cDNA-AFLP technique to compare gene expression profiles of a salt tolerant and a salt-sensitive cultivar of foxtail millet (Seteria italica) in response to salt stress to identify early responsive differentially expressed transcripts accumulated upon salt stress and validate the obtained result through quantitative real-time PCR (qRT-PCR). The expression profile was compared between a salt tolerant (Prasad) and susceptible variety (Lepakshi) of foxtail millet in both control condition (L0 and P0) and after 1 h (L1 and P1) of salt stress. We identified 90 transcript-derived fragments (TDFs) that are differentially expressed, out of which 86 TDFs were classified on the basis of their either complete presence or absence (qualitative variants) and 4 on differential expression pattern levels (quantitative variants) in the two varieties. Finally, we identified 27 non-redundant differentially expressed cDNAs that are unique to salt tolerant variety which represent different groups of genes involved in metabolism, cellular transport, cell signaling, transcriptional regulation, mRNA splicing, seed development and storage, etc. The expression patterns of seven out of nine such genes showed a significant increase of differential expression in tolerant variety after 1 h of salt stress in comparison to salt-sensitive variety as analyzed by qRT-PCR. The direct and indirect relationship of identified TDFs with salinity tolerance mechanism is discussed.




Similar content being viewed by others
References
Siminovitch, D., & Cloutier, Y. (1982). Twenty-four-hour induction of freezing and drought tolerance in plumules of winter rye seedlings by desiccation stress at room temperature in the dark. Plant Physiology, 60, 250–255.
Lang, V., Mantyla, E., Welin, B., Sundberg, B., & Tapio Palva, E. (1994). Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana abscisic acid. Plant Physiology, 104, 1341–1349.
Mantyla, E., Lang, V., & Tapio Palva, E. (1995). Role of abscisic acid in freezing tolerance, cold acclimation, and accumulation of LTI78 and RAB18 proteins in Arabidopsis thaliana. Plant Physiology, 107, 141–148.
Knight, H., Brandt, S., & Knight, M. R. (1998). A history of stress alters drought calcium signaling pathways in Arabidopsis. The Plant Journal, 16, 681–687. doi:10.1046/j.1365-313x.1998.00332.x.
Bray, E. A. (1997). Plant responses to water deficit. Trends in Plant Science, 2, 48–54. doi:10.1016/S1360-1385(97)82562-9.
Shinozaki, K., & Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology, 3, 217–223.
Kramer, A. (1996). Structure and function of proteins involved in mammalian pre-mRNA splicing. Annual Review of Biochemistry, 65, 367–409. doi:10.1146/annurev.bi.65.070196.002055.
Bohnert, H. J., & Jensen, R. G. (1996). Strategies for engineering water-stress tolerance in plants. Trends in Biotechnology, 14, 89–97. doi:10.1016/0167-7799(96)80929-2.
Hasegawa, P. M., Bressan, R. A., Zhu, J.-K., & Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology, 51, 463–499. doi:10.1146/annurev.arplant.51.1.463.
Xiong, L., Shumaker, K. S., & Zhu, J. K. (2002). Cell signaling during cold, drought and salt stresses. The Plant Cell, 14, S165–S183. doi:10.1105/tpc.010278.
Breyne, P., & Zabeau, M. (2001). Genome-wide expression analysis of plant cell cycle modulated genes. Current Opinion in Plant Biology, 4, 136–142. doi:10.1016/S1369-5266(00)00149-7.
Breyne, P., Dreesen, R., Cannoot, B., Rombaut, D., Vandepoele, K., Rombauts, S., et al. (2003). Quantitative cDNA-AFLP analysis for genome-wide expression studies. Molecular Genetics and Genomics, 269, 173–179.
Fukumura, R., Takahashi, H., Saito, T., Tsutsumi, T., Fujimori, A., Sato, S., et al. (2003). A sensitive transcriptome analysis method that can detect unknown transcripts. Nucleic Acids Research, 31, e94. doi:10.1093/nar/gng094.
Marathee, J. P. (1993). Structure and characteristics of the world millet economy. In K. W. Riley, S. C. Gupta, A. Seetharam, & J. N. Mushonga (Eds.), Advances in small millets (pp. 159–178). New Delhi: Oxford & IBH.
Sivaraman, L., & Ranjekar, P. K. (1984). Novel molecular features of millet genomes. Indian Journal of Biochemistry & Biophysics, 21, 299–303.
Sivaraman, L., & Gupta, V. S. (1986). DNA sequence organization in the genomes of three related millet plant species. Plant Molecular Biology, 6, 375–388. doi:10.1007/BF00027131.
Sanger, F., Nicklen, S., & Coulsen, A. R. (1977). DNA sequencing with chain terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America, 74, 5463–5467. doi:10.1073/pnas.74.12.5463.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402. doi:10.1093/nar/25.17.3389.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCt method. Methods (San Diego, Calif.), 25, 402–408. doi:10.1006/meth.2001.1262.
Vos, P., Hogers, R., Bleeker, M., Rijans, M., Van der Lee, T., Hornes, M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research, 23, 4407–4414. doi:10.1093/nar/23.21.4407.
Bachem, C. W. B., Van der Hoeven, R. S., de Bruijin, S. M., Vreugdenhil, D., Zabeau, M., & Visser, R. G. F. (1996). Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: Analysis of gene expression during tuber development. The Plant Journal, 9, 745–753. doi:10.1046/j.1365-313X.1996.9050745.x.
Baisakh, N., Subudhi, P. K., & Parami, N. P. (2006). cDNA-AFLP analysis reveals differential gene expression in response to salt stress in a halophyte Spartina alterniflora Loisel. Plant Science, 170, 1141–1149. doi:10.1016/j.plantsci.2006.02.001.
Tran, L. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a Drought-Responsive cis-Element in the early responsive to dehydration stress 1 Promoter. The Plant Cell, 16, 2481–2498. doi:10.1105/tpc.104.022699.
He, X. J., Mu, R. L., Cao, W. H., Zhang, Z. G., Zhang, J. S., & Chen, S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. The Plant Journal, 44, 903–916. doi:10.1111/j.1365-313X.2005.02575.x.
Smith, C. W. J., Patton, J. G., & Nadal-Ginard, B. (1989). Alternative splicing in the control of gene expression. Annual Review of Genetics, 23, 527–577. doi:10.1146/annurev.ge.23.120189.002523.
Sairam, R. K., & Tyagi, A. (2004). Physiology and molecular biology of salinity stress tolerance in plants. Current Science, 86, 407–421.
Bartels, D., & Sunkar, R. (2005). Drought and salt tolerance in plants. Critical Reviews in Plant Sciences, 24, 23–58. doi:10.1080/07352680590910410.
Krude, T., & Keller, C. (2001). Chromatin assembly during S phase: Contributions from histone deposition, DNA replication and the cell division cycle. Cellular and Molecular Life Sciences, 58, 665–672. doi:10.1007/PL00000890.
Ito, T., Ikehara, T., Nakagawa, T., Kraus, W. L., & Muramatsu, M. (2000). p300-Mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes & Development, 14, 1899–1907.
Krude, T. (1999). Chromatin assembly during DNA replication in somatic cells. European Journal of Biochemistry, 263, 1–5. doi:10.1046/j.1432-1327.1999.00508.x.
Ridgway, P., & Almouzni, G. (2001). Chromatin assembly and organization. Journal of Cell Science, 114, 2711–2712.
Green, C. M., & Almouzni, G. (2002). When repair meets chromatin—First in series on chromatin dynamics. EMBO Reports, 3, 28–33. doi:10.1093/embo-reports/kvf005.
Maser, R. S., & DePinho, R. A. (2002). Connecting chromosomes, crisis, and cancer. Science, 297, 565–569. doi:10.1126/science.297.5581.565.
Haushalter, K. A., & Kadonaga, J. T. (2003). Chromatin assembly by DNA-translocating motors. Nature Reviews. Molecular Cell Biology, 4, 613–620. doi:10.1038/nrm1177.
Abler, M. L., & Green, P. J. (1996). Control of mRNA stability in higher plants. Plant Molecular Biology, 32, 63–78. doi:10.1007/BF00039377.
Carrington, J. C., & Ambros, V. (2003). Role of microRNAs in plant and animal development. Science, 301, 336–338. doi:10.1126/science.1085242.
Kuhn, J. M., & Schroeder, J. I. (2003). Impacts of altered RNA metabolism on abscisic acid signaling. Current Opinion in Plant Biology, 6, 463–469. doi:10.1016/S1369-5266(03)00084-0.
Shi, H. Z., Lee, B.-H., Wu, S. J., & Zhu, J.-K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nature Biotechnology, 21, 81–85. doi:10.1038/nbt766.
Gong, Z. Z., Lee, H., Xiong, L., Jagendorf, A., Stevenson, B., & Zhu, J.-K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proceedings of the National Academy of Sciences of the United States of America, 99, 11507–11512. doi:10.1073/pnas.172399299.
Goeres, D. C., Van Norman, J. M., Zhang, W., Fauver, N. A., Spencer, M. L., & Sieburth, L. E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. The Plant Cell, 19, 1549–1564. doi:10.1105/tpc.106.047621.
Muhlrad, D., Decker, C. J., & Parker, R. (1994). Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 50>30 digestion of the transcript. Genes & Development, 8, 855–866. doi:10.1101/gad.8.7.855.
van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., & Seraphin, B. (2002). Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. The EMBO Journal, 21, 6915–6924. doi:10.1093/emboj/cdf678.
Xu, J., Yang, J. Y., Niu, Q. W., & Chua, N. H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. The Plant Cell, 18, 3386–3398. doi:10.1105/tpc.106.047605.
McGarry, R. C., Barron, Y. D., Carvalho, M. F., Hill, J. E., Gold, D., Cheung, E., et al. (2003). A novel Arabidopsis acetyltransferase interacts with the Geminivirus movement protein NSP. The Plant Cell, 15, 1605–1618. doi:10.1105/tpc.012120.
Chen, Y. W., Shao, G. H., & Chang, R. Z. (1997). The effect of salt stress on superoxide dismutase in various organelles of cotyledons of soybean seedlings. Acta Agronomica Sinica, 23, 214–219.
Gueta-Dahan, Y., Yaniv, Z., Zilinskas, B. A., & Ben-Hayyin, G. (1997). Salt and oxidative stress: Similar and specific response and their relation to salt tolerance in citrus. Planta, 203, 460–469. doi:10.1007/s004250050215.
Gomez, J. M., Hernandez, J. A., Jimenez, A., del Rio, L. A., & Sevilla, F. (1999). Differential response of antioxidative systems of chloroplasts and mitochondria to long term NaCl stress of pea plant. Free Radical Research, 31(Suppl.), 11–18. doi:10.1080/10715769900301261.
Sreenivasulu, N., Grimm, B., Wobus, U., & Weschke, W. (2000). Differential response of antioxidant components to salinity stress in salt-tolerant and salt-sensitive seedlings of foxtail millet (Setaria italica). Physiologia Plantarum, 109, 435–442. doi:10.1034/j.1399-3054.2000.100410.x.
Hernandez, J. A., Campillo, A., Jimenez, A., Alarcon, J. J., & Sevilla, F. (1999). Response of antioxidant systems and leaf water relations to NaCl stress in pea plants. The New Phytologist, 141, 241–251. doi:10.1046/j.1469-8137.1999.00341.x.
Hernandez, J. A., Jimerez, A., Mullineaux, P., & Sevilla, F. (2000). Tolerance of pea (Pisum sativum) to long term salt stress is associated with induction of antioxidant defences. Plant Cell Biology, 23, 853–862.
Gomez, G. M., Jimenez, A., Olmos, E., & Sevilla, F. (2004). Location and effects of long term NaCl stress on Superoxide dismutase and ascorbate peroxidase isoenzymes of pea (Pisum sativum cv Puget) chloroplasts. Journal of Experimental Botany, 55, 119–130. doi:10.1093/jxb/erh013.
Miller, G., Suzuki, N., Rizhsky, L., Hegie, A., Koussevitzky, S., & Mittler, R. (2007). Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiology, 144, 1777–1785. doi:10.1104/pp.107.101436.
Mathur, P. B., Vadez, V., & Sharma, K. K. (2008). Transgenic approaches for abiotic stress tolerance in plants: Retrospect and prospects. Plant Cell Reports, 27, 411–424. doi:10.1007/s00299-007-0474-9.
Kawasaki, S., Borchert, C., Deyholos, M., Wang, H., Brazille, S., Kawai, K., et al. (2001). Gene expression profiles during the initial phase of salt stress in rice. The Plant Cell, 13, 889–905.
Urao, T., Katagiri, T., Mizoguchi, T., Yamaguchi-Shinozaki, K., Hayashida, N., & Shinozaki, K. (1994). Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Molecular & General Genetics, 244, 331–340. doi:10.1007/BF00286684.
Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature, 406, 731–734. doi:10.1038/35021067.
Tähtiharju, S., Sangwan, V., Monroy, A. F., Dhindsa, R. S., & Borg, M. (1997). The induction of kin genes in cold-acclimating Arabidopsis thaliana. Evidence of a role for calcium. Planta, 203, 442–447. doi:10.1007/s004250050212.
Hwang, I., Sze, H., & Harper, J. F. (2000). A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 97, 6224–6229. doi:10.1073/pnas.97.11.6224.
Cheng, H.-S., Wilmann, R. M., Chen, H.-C., & Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium dependant protein kinase gene family. Plant Physiology, 129, 469–485. doi:10.1104/pp.005645.
Ludwig, A. A., Saitoh, H., Felix, G., Freymark, G., Miersch, O., Wasternack, C., et al. (2005). Ethylene mediated cross talk between calcium dependant protein kinase and MAPK signaling controls stress response in plants. Proceedings of the National Academy of Sciences of the United States of America, 102, 10736–10741. doi:10.1073/pnas.0502954102.
Binyamin, L., Falah, M., Portnoy, V., Soudry, E., & Gepstein, S. (2001). The early light induced protein is also produced during leaf senescence of Nicotiana tabacum. Planta, 212, 591–597. doi:10.1007/s004250000423.
Allakhverdiev, S. I., Nishiyama, Y., Miyairi, S., Yamamoto, H., Inagaki, M., Kanesaki, Y., et al. (2002). Salt stress inhibits the repair of photodamaged photosystem II by suppessing the transcription and translation of psbA genes in Synechocystis. Plant Physiology, 130, 1443–1453. doi:10.1104/pp.011114.
Olsen, A. N., Ernst, H. A., Lo Leggio, L., & Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends in Plant Science, 10, 79–87. doi:10.1016/j.tplants.2004.12.010.
Hegedus, D., Yu, M., Baldwin, D., Gruber, M., Sharpe, A., Parkin, I., et al. (2003). Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Molecular Biology, 53, 383–397. doi:10.1023/B:PLAN.0000006944.61384.11.
Guo, H. S., Xie, Q., Fie, J. F., & Chua, N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. The Plant Cell, 17, 1376–1386. doi:10.1105/tpc.105.030841.
Ouyang, B., Yang, T., Li, H., Zhang, L., Zhang, Y., Zhang, J., et al. (2007). Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and Microarray analysis. Journal of Experimental Botany, 58, 507–520. doi:10.1093/jxb/erl258.
Simpson, G. G., & Filipowicz, W. (1996). Splicing of precursors to mRNA in higher plants: Mechanism, regulation and sub-nuclear organization of the spliceosomal machinery. Plant Molecular Biology, 32, 1–41. doi:10.1007/BF00039375.
Brown, J. W. S., & Simpson, C. G. (1998). Splice site selection in plant pre-mRNA splicing. Annual Review of Plant Physiology and Plant Molecular Biology, 49, 77–95. doi:10.1146/annurev.arplant.49.1.77.
Lorkovic, Z. J., Wieczorek Kirk, D. A., Lambermon, M. H., & Filipowicz, W. (2000). Pre-mRNA splicing in higher plants. Trends in Plant Science, 5, 160–167. doi:10.1016/S1360-1385(00)01595-8.
Reddy, A. S. N. (2001). Nuclear pre mRNA splicing in plants. Critical Reviews in Plant Sciences, 20, 523–572. doi:10.1016/S0735-2689(01)80004-6.
Manley, J. L., & Tacke, R. (1996). SR proteins and splicing control. Genes & Development, 10, 1569–1579. doi:10.1101/gad.10.13.1569.
Sharp, P. A. (1994). Split genes and RNA splicing. Cell, 77, 805–815. doi:10.1016/0092-8674(94)90130-9.
Rodriguez, M., Canales, E., & Borras-Hidalgo, O. (2005). Molecular aspects of abiotic stress in plants. Biotecnologia Aplicada, 22, 1–10.
Acknowledgments
The authors thank the Director, National Institute of Plant Genome Research (NIPGR), for providing facilities and the Department of Biotechnology, Government of India, for providing financial support. Acknowledgement is also due toward Dr. Debasis Chattopadhyay (NIPGR, New Delhi) for his review and suggestions.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Jayaraman and S. Puranik contributed equally to this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jayaraman, A., Puranik, S., Rai, N.K. et al. cDNA-AFLP Analysis Reveals Differential Gene Expression in Response to Salt Stress in Foxtail Millet (Setaria italica L.). Mol Biotechnol 40, 241–251 (2008). https://doi.org/10.1007/s12033-008-9081-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-008-9081-4