Abstract
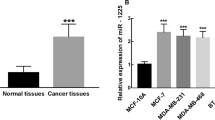
Dysregulation of microRNA plays critical roles in various malignancies. However, whether the aberrant expression of miR-215 in breast cancer is associated with malignancy, metastasis, or prognosis remains unknown. In this study, we demonstrated that the relative level of miR-215 expression was lower in cancer tissues compared with adjacent non-malignant tissues (p < 0.001). Low-miR-215 expression was observed to be closely correlated with higher tumor grade (p = 0.008), human epidermal growth factor receptor 2 (HER2) positivity (p = 0.006), HER2 positive breast cancer subtype (p = 0.016), and lymph node metastasis (p = 0.039). Moreover, patients with low-miR-215 expression showed shorter 5-year disease-specific survival (DSS) than the high-miR-215 expression group (p = 0.007). Multivariate analysis results revealed that miR-215 downexpression was an unfavorable prognostic factor for DSS in addition to tumor size, ER, and lymph node metastasis. Our results support the potential of miR-215 as a prognostic predictor for breast cancer with its high expression in cancer tissues and its relationship with other clinicopathologic factors and survival.



Similar content being viewed by others
References
Motamedolshariati M, Memar B, Aliakbaian M, Shakeri MT, Samadi M, Jangjoo A. Accuracy of prognostic and predictive markers in core needle breast biopsies compared with excisional specimens. Breast Care. 2014;9(2):107–10. doi:10.1159/000360787.
Zhang BN, Cao XC, Chen JY, Chen J, Fu L, Hu XC, et al. Guidelines on the diagnosis and treatment of breast cancer (2011 edition). Gland Surg. 2012;1(1):39–61. doi:10.3978/j.issn.2227-684X.2012.04.07.
Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–89. doi:10.1016/S1470-2045(13)70567-9.
Rutnam ZJ, Yang BB. The involvement of microRNAs in malignant transformation. Histol Histopathol. 2012;27(10):1263–70.
Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–58. doi:10.1016/S1470-2045(12)70073-6.
Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi:10.1146/annurev-pathol-012513-104715.
Mulrane L, McGee SF, Gallagher WM, O’Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73(22):6554–62. doi:10.1158/0008-5472.CAN-13-1841.
Liu H. MicroRNAs in breast cancer initiation and progression. Cell Mol Life Sci: CMLS. 2012;69(21):3587–99. doi:10.1007/s00018-012-1128-9.
Pena-Chilet M, Martinez MT, Perez-Fidalgo JA, Peiro-Chova L, Oltra SS, Tormo E, et al. MicroRNA profile in very young women with breast cancer. BMC Cancer. 2014;14(1):529. doi:10.1186/1471-2407-14-529.
Bischoff A, Huck B, Keller B, Strotbek M, Schmid S, Boerries M, et al. miR-149 functions as a tumor suppressor by controlling breast epithelial cell migration and invasion. Cancer Res. 2014;. doi:10.1158/0008-5472.CAN-13-3319.
Schrauder MG, Strick R, Schulz-Wendtland R, Strissel PL, Kahmann L, Loehberg CR, et al. Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One. 2012;7(1):e29770. doi:10.1371/journal.pone.0029770.
Fu SW, Chen L, Man YG. miRNA biomarkers in breast cancer detection and management. J Cancer. 2011;2:116–22.
Perez-Rivas LG, Jerez JM, Carmona R, de Luque V, Vicioso L, Claros MG, et al. A microRNA signature associated with early recurrence in breast cancer. PLoS One. 2014;9(3):e91884. doi:10.1371/journal.pone.0091884.
Smeets A, Daemen A, Vanden Bempt I, Gevaert O, Claes B, Wildiers H, et al. Prediction of lymph node involvement in breast cancer from primary tumor tissue using gene expression profiling and miRNAs. Breast Cancer Res Treat. 2011;129(3):767–76. doi:10.1007/s10549-010-1265-5.
Tanic M, Yanowski K, Gomez-Lopez G, Socorro Rodriguez-Pinilla M, Marquez-Rodas I, Osorio A, et al. microRNA expression signatures for the prediction of BRCA1/2 mutation-associated hereditary breast cancer in paraffin-embedded formalin-fixed breast tumors. Int J Cancer. 2014;. doi:10.1002/ijc.29021.
Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68(24):10105–12. doi:10.1158/0008-5472.CAN-08-1846.
Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18(4):367–81. doi:10.1016/j.ccr.2010.09.005.
Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, et al. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16(11):2655–66. doi:10.1111/j.1582-4934.2012.01579.x.
Karaayvaz M, Pal T, Song B, Zhang C, Georgakopoulos P, Mehmood S, et al. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. 2011;10(4):340–7. doi:10.1016/j.clcc.2011.06.002.
van Schooneveld E, Wouters MC, Van der Auwera I, Peeters DJ, Wildiers H, Van Dam PA, et al. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res: BCR. 2012;14(1):R34. doi:10.1186/bcr3127.
Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–55. doi:10.1038/nature10888.
Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497(7449):378–82. doi:10.1038/nature12108.
Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ, et al. MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97(6):853–61. doi:10.1002/bjs.7000.
White NM, Khella HW, Grigull J, Adzovic S, Youssef YM, Honey RJ, et al. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer. 2011;105(11):1741–9. doi:10.1038/bjc.2011.401.
Necela BM, Carr JM, Asmann YW, Thompson EA. Differential expression of microRNAs in tumors from chronically inflamed or genetic (APC(Min/+)) models of colon cancer. PLoS One. 2011;6(4):e18501. doi:10.1371/journal.pone.0018501.
Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103(24):9136–41. doi:10.1073/pnas.0508889103.
Deng Y, Huang Z, Xu Y, Jin J, Zhuo W, Zhang C, et al. MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett. 2014;342(1):27–35. doi:10.1016/j.canlet.2013.08.033.
Liu F, You X, Chi X, Wang T, Ye L, Niu J, et al. Hepatitis B virus X protein mutant HBxDelta127 promotes proliferation of hepatoma cells through up-regulating miR-215 targeting PTPRT. Biochem Biophys Res Commun. 2014;444(2):128–34. doi:10.1016/j.bbrc.2014.01.004.
Zheng L, Ren JQ, Li H, Kong ZL, Zhu HG. Downregulation of wild-type p53 protein by HER-2/neu mediated PI3 K pathway activation in human breast cancer cells: its effect on cell proliferation and implication for therapy. Cell Res. 2004;14(6):497–506. doi:10.1038/sj.cr.7290253.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors Shu-wei Zhou and Bei-bei Su contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhou, Sw., Su, Bb., Zhou, Y. et al. Aberrant miR-215 expression is associated with clinical outcome in breast cancer patients. Med Oncol 31, 259 (2014). https://doi.org/10.1007/s12032-014-0259-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0259-2