Abstract
Improved whole brain angiographic and velocity-sensitive MRI is pushing the boundaries of noninvasively obtained cerebral vascular flow information. The complexity of the information contained in such datasets calls for automated algorithms and pipelines, thus reducing the need of manual analyses by trained radiologists. The objective of this work was to lay the foundation for such automated pipelining by constructing and evaluating a probabilistic atlas describing the shape and location of the major cerebral arteries. Specifically, we investigated how the implementation of a non-linear normalization into Montreal Neurological Institute (MNI) space improved the alignment of individual arterial branches. In a population-based cohort of 167 subjects, age 64–68 years, we performed 4D flow MRI with whole brain volumetric coverage, yielding both angiographic and anatomical data. For each subject, sixteen cerebral arteries were manually labeled to construct the atlas. Angiographic data were normalized to MNI space using both rigid-body and non-linear transformations obtained from anatomical images. The alignment of arterial branches was significantly improved by the non-linear normalization (p < 0.001). Validation of the atlas was based on its applicability in automatic arterial labeling. A leave-one-out validation scheme revealed a labeling accuracy of 96 %. Arterial labeling was also performed in a separate clinical sample (n = 10) with an accuracy of 92.5 %. In conclusion, using non-linear spatial normalization we constructed an artery-specific probabilistic atlas, useful for cerebral arterial labeling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cerebrovascular imaging is critical in diagnosing several neurological disorders (Kronzon and Tunick 2006; Mueller et al. 2005) as well as in research investigating cerebrovascular physiology and pathophysiology (Amin-Hanjani et al. 2015; Muller and Van Der Graaf 2012; Rivera-Rivera et al. 2015; Zarrinkoob et al. 2015). Several recent technological advancements, embodied in 4D flow MRI, allow for acquisition of data such as velocity, flow rate, turbulence patterns and pulsatility in brain arteries (Frydrychowicz et al. 2011). Therefore, post-processing methods that can transform these datasets into meaningful standardised quantitative descriptions of blood flow in the individual will soon be needed.
Probabilistic tissue maps are commonly used for tissue segmentation in the whole brain (Ashburner and Friston 2005) and in specific areas such as the cerebellum (Diedrichsen et al. 2009; van Baarsen et al. 2016) in both humans and animals (Love et al. 2016). However, such methods have not been explored for labeling and segmentation of cerebral arterial branches.
An atlas describing the spatial distribution of individual arteries could enable a high degree of automation in applications that require labeling of vascular segments. Pioneering work has provided detailed descriptions on individual segments (Nowinski et al. 2009a, b, 2013), as well as probabilistic information on the morphology of the cerebrovascular tree (Forkert et al. 2013; Mut et al. 2014). Recently, we combined these two properties into a probabilistic artery-specific atlas and an automatic, atlas-based artery-identification method (AAIM) (Dunås et al. 2016). The promising results from that proof-of-concept study motivate the construction of an atlas based on a large population-based sample, which would give a more representative and improved spatial coverage of the anatomic variation between individuals.
To validate such an arterial atlas in a meaningful way, the purpose of the atlas must be considered. Therefore, atlases developed for segmentation and labeling are generally evaluated for that specific task (Dunås et al. 2016; Forkert et al. 2013; Passat et al. 2005). Another important evaluation is based on describing the underlying spatial alignment of the structures that are included in the atlas.
The aim of this study was to create a stereotactic and probabilistic cerebral arterial atlas by manually labeling 167 high-resolution 4D flow MRI angiographic scans. This atlas was validated based on its applicability for arterial labeling, and the impact of the normalization process was investigated by comparing non-linear normalization with rigid-body alignment.
Materials and Methods
In this study, 2360 cerebral arteries were manually labeled in 4D flow MRI from 167 healthy elderly subjects and used to create the atlas. The workflow had five main steps: 1. Data acquisition; 2. Spatial image normalization and preprocessing; 3. Atlas construction; 4. Comparison to rigid-body alignment; and 5. Atlas validation.
1. Data Acquisition
Subjects from the Population
COBRA (Cognition, Brain and Aging) (Nevalainen et al. 2015) is a large, population-based, prospective MR imaging study. In summary, subjects between 64 and 68 years of age were randomly selected from the population registry of Umeå, Sweden. Subjects with medical conditions or medical or surgical interventions that could alter brain function or cognitive performance such as history of brain trauma or stroke, dementia, diabetes, functional impairment or movement disorders (e.g., Parkinson’s disease), epilepsy, intellectual disability, psychological disorders, and ongoing malignancy treatment, were excluded, as well as subjects with contraindications for MRI.
As a part of the COBRA study, we collected 4D flow MRI data from 181 subjects (age 66.2 ± 1.2, M = 100, F = 81). Out of these, thirteen subjects were excluded due to constraints regarding data quality (e.g. motion artifacts) and one due to a vascular malformation. This resulted in a sample of 167 subjects (age 65.8 ± 1.2, M = 97, F = 70) on which the atlas was based.
Clinical Sample
For a pilot test on a clinical sample, ten patients with transient ischemic attacks (n = 6) or lacunar infarcts (n = 4) were included. This diagnosis was based on case history, neurological and brain MRI examination. All patients were also investigated with CT angiography (0.6 mm slices). CT did not reveal any stenosis or occlusion of internal carotid, vertebral or basilar arteries, or in the middle, anterior or posterior cerebral arteries.
MRI
The MRI data used in this study were collected on a 3 Tesla scanner (Discovery MR 750; GE Healthcare, Milwaukee, WI, USA) with a 32-channel head coil, using a balanced 5-point 4D flow MRI (Johnson and Markl 2010) covering the intracranial cavity. The scan time for the 4D flow sequence was approximately nine minutes. Imaging parameters were: velocity encoding, 110 cm/s; TR/TE, 6.5/2.7 ms; flip angle, 8°; bandwidth, 166.67 kHz; radial projections, 1600; acquisition resolution, 300 × 300 × 300; imaging volume, 220 × 220 × 220 mm; reconstruction matrix size, 320 × 320 × 320 (zero padded interpolation); and voxel size 0.7 × 0.7 × 0.7 mm3. From the 4D flow MRI, a structural magnitude image (MT1w) and an angiographic image (AI) were reconstructed (Dunås et al. 2016).
2. Image Processing
Normalization
Tissue probability maps for white matter, grey matter and cerebrospinal fluid were generated from the structural MT1w using SPM8’s New Segment. From these tissue probability maps, a study-specific brain template was generated using SPM8’s DARTEL (Ashburner 2007). In that process, a subject-specific transformation field that described the nonlinear transformation from each subject to the study template was also generated. With these transformation fields, the AI of each subject was transformed to match the group template and then normalized to stereotactic Montreal Neurological Institute (MNI) space by aligning it with the MNI152-template (Evans et al. 2012) using an affine transformation.
Vascular Segmentation and Skeleton Construction
The AI was smoothed with a low-pass box filter with a kernel size of three voxels. To give complete vessel coverage without including neighboring static tissue (Wåhlin et al. 2012), the image was binarized by thresholding at 18 % of the maximum intensity value.
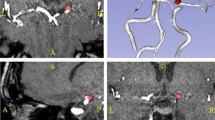
To separate the branches of the vascular tree a vascular skeleton was extracted from the binary image using an automatic method containing three steps: 1. The binary vessel tree was gradually thinned until a one-voxel thick skeleton was obtained (Palàgyi and Kuba 1998); 2. The vascular skeleton was pruned to remove loops and branches shorter than eight voxels; 3. The vascular skeleton was divided into branches separated by junction-points, and each branch was assigned an identification number (Chen and Molloi 2003). Figure 1 illustrates the segmentation and skeleton construction.
3. Atlas Construction
Included Arteries
The arteries included in the atlas were selected based on their fundamental role in the cerebral arterial circulation. The included arteries were: left and right internal carotid artery (ICA); basilar artery (BA); left and right vertebral artery (VA); left and right posterior cerebral artery (PCA); left and right, proximal and distal middle cerebral artery (MCA); left, right and distal anterior cerebral artery (ACA); and left and right posterior communicating artery (PCoA).
Manual Arterial Labeling
The arteries were manually labeled using an in-house tool developed in Matlab (Mathworks, MA, USA). The vessels of the brain were visualized as a rotatable 3D volume. The arterial segments forming each artery were selected, and the corresponding vascular skeleton branches were labeled and saved. The border between two arteries was primarily defined according to junction points in the vascular skeleton. When no such junction points were present (e.g. due to an absent branch) the border was manually determined based on morphology such as changes in diameter or direction (Osborn 1999).
To re-inflate the labeled arteries, the vascular skeleton branches with the corresponding labels were dilated with a kernel of size seven voxels, and the resulting volume was multiplied with the binary vessel tree. The labeled arteries were then visually inspected and approved by a neurologist (LZ). When there was uncertainty regarding which branches that should be included (primarily in MCA), a consensus decision was made by LZ and TD (5 and 3 years of experience in neurovascular anatomy). An example of manually labeled arteries from one subject is presented in Fig. 2.
An example of manually labeled arteries for one subject. For ACAdistal, all main branches distal to the anterior communicating artery were selected (1–3 branches of the pericallosal artery depending on morphology, A2–A3 level). PCA was cropped at P3 level, distal to pons, to get a uniform length (Osborn 1999). MCA was divided into a proximal (MCA) and a distal (MCAdistal) part. The proximal part consists of the M1 segment, pre- and post-bifurcation. The MCAdistal includes the full visible length of MCA, or until it reaches the cortex (M2 and M3 segments). The border between the proximal and distal part was set at the genu where the MCA takes a turn in the posterior direction (Osborn 1999). Only the branches that extend posteriorly (M2) and laterally (M3) were included. For M1, branches forming/preceding the main M2 branches, or having the same direction as those doing so, were included. The direction and continuity of the arteries were decided by visual inspection. Since MCAdistal consists of several branches, the individual variation at M3 level was too large for it to be useful to construct a separate probability map. Note that in the vascular segmentation process, gaps sometimes arise in low-flow arteries, here seen in the MCAdistal on the left side of the figure
A probability map was constructed for each artery by adding together the binary volumes of the re-inflated arteries for all subjects and dividing the value of each voxel by the number of included arteries. This resulted in a 3D volume with values between zero and one, corresponding to the proportion of included arteries that overlapped in each voxel. These 16 probability maps together form the atlas, which was denoted Umeå Brain Arteries (UBA167).
4. Impact of Normalization
Rigid-Body Atlas
To determine the contribution from the DARTEL normalization, the UBA167 was compared with an atlas where the vascular trees were aligned using a rigid-body transform. The obtained volumes were transformed to native space and aligned to the MNI152-template using a rigid-body transform in SPM8. This procedure was required since the manual labeling of each subject was performed after DARTEL normalization. Probability maps for each artery were calculated in the same way as for UBA167.
Since the arteries were extracted after DARTEL normalization, the arterial volumes had to be transformed back to native space before aligning them to the MNI-template in order to create the rigid-body atlas. Due to partial volume effects, the images had to be re-binarized after the rigid-body alignment. This was done using a threshold adapted for volume conservation of each probability map.
Atlas Characterization
The normalization procedure was evaluated by comparing the UBA167 to the rigid-body atlas. Four measurements were used to describe the spatial distribution of arteries in terms of alignment between subjects and separation between arteries in the probability maps. For each artery probability map, the measurements were: 1. Concatenated volume calculated from the number of non-zero voxels, i.e. voxels occupied by an artery in any of the 167 subjects; 2. The arterial volume ratio (AVR) obtained by dividing the concatenated volume of the probability map by the average arterial volume across the included subjects; 3. The dominating volume of each probability map calculated as the percentage of voxels where the artery in question had higher probability than any other overlapping artery; 4. The maximum value of each probability map.
A low AVR equals a high spatial agreement between subjects, and hence a high specificity for separating arteries from background. The dominating volume describes how well the probability maps are separated, which could be translated to the specificity for separating different arteries.
5. Atlas Validation
Leave-One-Out Validation
To evaluate the sensitivity of the UBA167, it had to be applied to a new sample of subjects. This was done using a leave-one-out approach where a target subject was removed from the UBA167 and the modified atlas based on the remaining 166 subjects was used to label the arteries of the target subject, using the previously described automatic identification method (AAIM) (Dunås et al. 2016). This process was repeated for all subjects in the cohort. The labeled images were approved or disapproved according to the following criteria:
-
ICA: 2 cm cervical segment
-
VA: 1 cm straight segment in conjunction to the foramen magnum
-
BA: 1 cm segment anywhere in the artery
-
PCA: 1 cm segment at P2–P3, i.e. distal to PCoA
-
MCA: 1 cm segment in M1
-
MCAdistal: 1 cm segment in the sylvian fissure
-
ACA: 5 mm segment in A1, proximal to the anterior communicating artery
-
ACAdistal: 1 cm segment distal to anterior communicating artery
-
PCoA: 5 mm segment anywhere in the artery
Agreement between automatic and manual labeling was described and divided into six categories:
-
Correctly identified existing: The artery was present in the manual reference and was localized.
-
Correctly identified nonexisting: The artery was not present in the manual reference and was marked as not found by the AAIM.
-
Mislabeled existing: The artery was present in the manual reference but at least a part of the labeled volume was inaccurate. A subjective decision in each case was used to determine whether the degree of mislabeling was clinically relevant.
-
Mislabeled nonexisting: The artery was not present in the manual reference but was marked as existing by the AAIM.
-
Not identified: The artery was not labeled, even though it did exist in the manual reference.
-
Too short: The identified segment belonged to the correct artery but did not include the required segment.
Clinical Validation
For the UBA167 to be clinically relevant, it has to be applicable to vascular diseases. As a proof-of-concept of this generalizability, the UBA167 was applied to a clinical sample of ten patients with transient ischemic attacks or lacunar infarcts. The labeling was performed with the AAIM. Again, the labeled images were evaluated using the previously mentioned criteria. In addition, results were compared to a reference obtained from computed tomography (CT) angiograms.
Results
The arterial atlas, UBA167, is presented in Fig. 3, and the specific contribution of applying non-linear image normalization over rigid-body co-registration is seen in Fig. 4.
Visualization of the probabilistic (a–c) and artery - specific (d–f) properties of the UBA167 shown in axial, coronal and sagittal view. The probability values (a–c) are visualized with a heat map, min = 0, mid = 0.1 and max = 1.0. In (d–f), each voxel is labeled as the artery with the highest probability
As revealed by the more confined probability maps, the non-linear image normalization generated a higher degree of arterial alignment than the rigid-body transformation.
Prevalence of Arteries
The number of arteries included in UBA167 and their average volumes are presented in Table 1. In total 2360 arteries were manually labeled. ICA, MCA and ACAdistal were present in all subjects. BA and PCA were present in over 98 % of the subjects. VA, ACA and MCAdistal were present in at least 90 % of subjects, while only 38 % (63/167) had a PCoA on any side, and 28.7 % (48/167) had a fetal PCoA (missing P1).
Arterial Volume Ratio
The AVR of each probability map is presented in Table 1 and reflects how well the normalization method works and to which extent each artery permits such normalization. A perfect normalization would result in an AVR of 1.0, and higher values indicate a less effective normalization. The AVR for the probability maps in UBA167 were significantly lower than for the rigid-body atlas (p < 0.001, Wilcoxon signed-rank test). When looking at the whole atlas, the AVR for the UBA167 was 13.7, and the corresponding value for the rigid-body atlas was 29.3.
In general, the proximal arteries of the anterior circulation (ICA, MCA, ACA and PCoA) had a low AVR (6.4 to 14.8) compared with posterior (PCA, VA and BA, AVR 19.0 to 26.1) and distal arteries (MCAdistal and ACAdistal, AVR 18.2 to 42.8). The AVR was negatively correlated with the accuracy of the leave-one-out validation (p = 0.035, Spearman correlation, rho = − 0.46, one-tailed probability).
Dominating Volume and Maximum Probability
The dominating volume of each probability map and the maximum probability value are also presented in Table 1. The average dominating volume for UBA167 was 85.8 %, compared with 74.9 % in the rigid-body atlas, and this difference was statistically significant (p < 0.005, Wilcoxon signed-rank test), indicating that the DARTEL normalization did indeed improve the spatial alignment between subjects.
The maximum probability value is also a measurement of spatial distribution. For UBA167, many arteries had voxels where over 80 % of subjects overlapped, ICA even had 100 % overlap in some voxels. For the rigid-body atlas, no more than 53 % of the subjects overlapped in any single voxel.
Correctly Identifying Cerebral Arteries
The result from the leave-one-out validation can be found in Table 2. The average labeling accuracy for the leave-one-out validation was 96 %. Lower values were observed for VA, PCoA and left MCAdistal. The specificity could not be calculated for all arteries because the number of true negatives was zero for most arteries.
The average labeling accuracy in the clinical sample was 92.5 % (Table 3). In two patients, the identified right PCA was too short to fulfill the evaluation criteria. This was also the case for the right VA in two patients. One ACAdistal and one MCAdistal on each side were not identified. The existing PCoAs were not identified in the clinical sample. In the CT-angiography that serves as the reference in this evaluation, PCoA was found bilaterally in one subject and unilaterally in three subjects.
Discussion
The probabilistic atlas Umeå Brain Arteries (UBA167), which contains 16 major brain arteries, was constructed based on manually labeled 4D flow MRI angiography from 167 subjects. This is the first project describing the “artery-specific” 3D spatial distribution within the human brain. UBA167 can be used to automatically label and segment the major cerebral arteries for future automated blood flow quantification. Similar to successful brain tissue atlases (Diedrichsen et al. 2009; Tzourio-Mazoyer et al. 2002; van Baarsen et al. 2016), UBA167 is provided in MNI space. We believe that multiple additional neuroimaging applications can be developed around this atlas. For example, the atlas can be used to extract geometrical measures such as size and tortuosity of brain arteries. In addition, the atlas can be used to detect and correct for signals in large arteries that can degrade the physiological interpretation of MRI data (e.g. arterial spin labeling, functional MRI, diffusion imaging). For example, in functional MRI, removal of vascular contamination is of known importance (Kiviniemi et al. 2003).
Achieving sufficient spatial inter-individual alignment of brain arteries cannot be taken for granted, as demonstrated by the fact that DARTEL normalization outperformed rigid-body transformation. This finding is analogous to results obtained for brain structural normalization (Klein et al. 2009). In the current paper we hypothesized that normalization prior to atlas labeling would provide a feasible pipeline for automated labeling. Our results supported this hypothesis by showing that all major arteries were labeled with high accuracy (>95 %), indicating that the achieved inter-individual spatial variability permitted efficient atlas construction.
UBA167 was constructed from a population-based sample (Nevalainen et al. 2015), and can thus be expected to represent a wide range of vascular morphology. This may have contributed to our finding of a high accuracy when using UBA167 on the ten stroke patients. An atlas based on a more selected sample (for instance only using subjects without hypertension, hyperlipidemia and obesity) could have resulted in less generalizability as such cardiovascular risk factors may affect the anatomy and tortuosity of cerebral vessels (Bullitt et al. 2009; Hiroki et al. 2002).
The atlas is developed from data on individuals from a limited age span. We still expect a high degree of generalizability of the atlas, assuming that future studies normalize data according to the present study. However, future studies using the atlas have to verify its functionality for that particular sample.
The main innovation of the UBA167 is the combination of the artery-specific probabilistic properties with the fully automatic labeling of the major cerebral arteries. The UBA167 is thus prepared for automatic blood flow quantification in 4D flow MRI data. Pioneering work on characterization of the cerebral arterial system has been based on small sample sizes (Nowinski et al. 2009a, 2011, 2013) or has not included specific probabilities for individual arterial segments (Forkert et al. 2013; Mut et al. 2014; Wright et al. 2013). Knowledge of vascular morphology has also been used for semi-automated segmentation and labeling (Bogunović et al. 2012; Ghanavati et al. 2014; Passat et al. 2006).
Our finding of a maximum probability value of 1.0 in the bilateral ICA means that there exist one or more voxels in each ICA where all 167 subjects overlapped. These voxels provide the compelling option to explore seeding points, combined with region-growing segmentation schemes, since the probability that those voxels will be within the ICA for a new subject is very high (Passat et al. 2005). UBA167 as a whole can also be used to specify the region of interest or provide spatial information for arterial segmentation to reduce computational time (Passat et al. 2006).
UBA167 enabled a high labeling accuracy (Tables 2 and 3). Although the input data for the labeling validation was from 4D flow MRI, a comparable imaging technique can be expected to produce equally accurate results when combined with the AAIM.
We used a velocity encoding of 110 cm/s for the 4D flow sequence. This value was selected to avoid aliasing in large arteries. This option is not optimal to visualize and quantify the slow-velocity blood flow of the posterior communicating cerebral arteries (Dunås et al. 2016). Indeed, this was clearly evident when the CT angiogram images from the patient group were reviewed. Here CT angiography detected additional PCoA arteries that were not visible on the thresholded AI. This is not a strict limitation of UBA167 and labeling procedure per se, but rather a manifestation of differences in the underlying measurement techniques. However, with respect to the development of an automatic flow assessment of 4D flow MRI data, the high labeling accuracy for arteries that had detectable velocities was very promising. Future developments could improve the labeling accuracy of VA segments by improving the labeling criteria. The evaluation conditions for the labeling were set to ensure that segments needed for future blood flow quantification were correctly identified, therefore some existing arteries could be marked as too short.
The UBA167 included the major cerebral arteries, therefore it does not allow investigation of more distal arteries. Such distal arteries could also be added, but the increasing inter-individual variations in anatomy at that depth in the arterial tree would potentially limit the usefulness of such expansions. This effect can be seen in the MCAdistal in Fig. 3, appearing with lower spatial alignment between subjects compared with more proximal arteries. Since the P1 and P2 segments were defined as the same artery, and the anterior communicating artery was not included, UBA167 cannot differentiate between some of the typical morphological variants. The standard variations that can be automatically identified are missing A1 or PCoA (Krabbe-Hartkamp et al. 1998).
Conclusion
UBA167 is an artery-specific probabilistic atlas based on 16 manually-labeled major cerebral arteries from 167 subjects. The UBA167 enables a high accuracy in automatic arterial labeling in both population-based subjects and in ischemic patients. Comparison to rigid-body alignment showed a large improvement in spatial alignment for non-linear normalization. Taken together, this study provides compelling first evidence for the usefulness of a probabilistic stereotactic atlas of the major cerebral arteries.
Information Sharing Statement
The atlas (RRID:SCR_016319) will be made publically available at time of publication at http://www.nitrc.org/projects/brainarteries
Abbreviations
- AAIM:
-
Automatic atlas based Artery Identification Method
- ICA:
-
Internal carotid artery
- VA:
-
Vertebral artery
- BA:
-
Basilar artery
- PCA:
-
Posterior cerebral artery
- MCA:
-
Middle cerebral artery
- ACA:
-
Anterior cerebral artery
- PCoA:
-
Posterior communicating artery
- AI:
-
Angiographic image
- AVR:
-
Arterial volume ratio
- MNI:
-
Montreal Neurological Institute
References
Amin-Hanjani, S., Du, X., Pandey, D. K., Thulborn, K. R., & Charbel, F. T. (2015). Effect of age and vascular anatomy on blood flow in major cerebral vessels. Journal of Cerebral Blood Flow & Metabolism, 35(2), 312–318. doi:10.1038/jcbfm.2014.203.
Ashburner, J. (2007). A fast diffeomorphic image registration algorithm. NeuroImage, 38(1), 95–113. doi:10.1016/j.neuroimage.2007.07.007.
Ashburner, J., & Friston, K. J. (2005). Unified segmentation. NeuroImage, 26(3), 839–851. doi:10.1016/j.neuroimage.2005.02.018.
Bogunović, H., Pozo, J. M., Cárdenes, R., Villa-Uriol, M. C., Blanc, R., Piotin, M., & Frangi, A. F. (2012). Automated landmarking and geometric characterization of the carotid siphon. Medical Image Analysis, 16(4), 889–903. doi:10.1016/j.media.2012.01.006.
Bullitt, E., Rahman, F. N., Smith, J. K., Kim, E., Zeng, D., Katz, L. M., & Marks, B. L. (2009). The effect of exercise on the cerebral vasculature of healthy aged subjects as visualized by MR angiography. American Journal of Neuroradiology, 30(10), 1857–1863. doi:10.3174/ajnr.A1695.
Chen, Z., & Molloi, S. (2003). Automatic 3D vascular tree construction in CT angiography. Computerized Medical Imaging and Graphics, 27(6), 469–479. doi:10.1016/S0895-6111(03)00039-9.
Diedrichsen, J., Balsters, J. H., Flavell, J., Cussans, E., & Ramnani, N. (2009). A probabilistic MR atlas of the human cerebellum. NeuroImage, 46(1), 39–46. doi:10.1016/j.neuroimage.2009.01.045.
Dunås, T., Wåhlin, A., Ambarki, K., Zarrinkoob, L., Birgander, R., Malm, J., & Eklund, A. (2016). Automatic labeling of cerebral arteries in magnetic resonance angiography. Magnetic Resonance Materials in Physics, Biology and Medicine, 29(1), 39–47. doi:10.1007/s10334-015-0512-5.
Evans, A. C., Janke, A. L., Collins, D. L., & Baillet, S. (2012). Brain templates and atlases. NeuroImage, 62(2), 911–922. doi:10.1016/j.neuroimage.2012.01.024.
Forkert, N. D., Fiehler, J., Suniaga, S., Wersching, H., Knecht, S., & Kemmling, A. (2013). A statistical cerebroarterial atlas derived from 700 MRA datasets. Methods of Information in Medicine, 52(6), 467–474. doi:10.3414/ME13-02-0001.
Frydrychowicz, A., François, C. J., & Turski, P. A. (2011). Four-dimensional phase contrast magnetic resonance angiography: potential clinical applications. European Journal of Radiology, 80(1), 24–35. doi:10.1016/j.ejrad.2011.01.094.
Ghanavati, S., Lerch, J. P., & Sled, J. G. (2014). Automatic anatomical labeling of the complete cerebral vasculature in mouse models. NeuroImage, 95, 117–128. doi:10.1016/j.neuroimage.2014.03.044.
Hiroki, M., Miyashita, K., & Oda, M. (2002). Tortuosity of the white matter medullary arterioles is related to the severity of hypertension. Cerebrovascular Diseases, 13, 242–250.
Johnson, K. M., & Markl, M. (2010). Improved SNR in phase contrast velocimetry with five-point balanced flow encoding. Magnetic Resonance in Medicine, 63(2), 349–355. doi:10.1002/mrm.22202.
Kiviniemi, V., Kantola, J. H., Jauhiainen, J., Hyvärinen, A., & Tervonen, O. (2003). Independent component analysis of nondeterministic fMRI signal sources. NeuroImage, 19(2), 253–260. doi:10.1016/S1053-8119(03)00097-1.
Klein, A., Andersson, J., Ardekani, B. a., Ashburner, J., Avants, B., Chiang, M. C., et al. (2009). Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage, 46(3), 786–802. doi:10.1016/j.neuroimage.2008.12.037.
Krabbe-Hartkamp, M. J., van der Grond, J., de Leeuw, F.-E., de Groot, J.-C., Algra, A., Hillen, B., et al. (1998). Circle of Willis: morphologic variation on three-dimensional time-of-flight MR angiograms. Radiology, 207(1), 103–111.
Kronzon, I., & Tunick, P. a. (2006). Aortic atherosclerotic disease and stroke. Circulation, 114(1), 63–75. doi:10.1161/CIRCULATIONAHA.105.593418.
Love, S. A., Marie, D., Roth, M., Lacoste, R., Nazarian, B., Bertello, A., et al. (2016). The average baboon brain: MRI templates and tissue probability maps from 89 individuals. NeuroImage, 13, 526–533. doi:10.1016/j.neuroimage.2016.03.018.
Mueller, S. G., Weiner, M. W., Thal, L. J., Petersen, R. C., Jack, C. R., Jagust, W., et al. (2005). Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s disease neuroimaging initiative (ADNI). Alzheimer’s & Dementia : the Journal of the Alzheimer’s Association, 1(1), 55–66. doi:10.1016/j.jalz.2005.06.003.
Muller, M., & Van Der Graaf, Y. (2012). Hypertension and longitudinal changes in cerebral blood flow: the SMART-MR study. Annals of Neurology, 71(6), 825–833. doi:10.1002/ana.23554.
Mut, F., Wright, S., Ascoli, G. A., & Cebral, J. R. (2014). Morphometric, geographic, and territorial characterization of brain arterial trees. International Journal of Numerical Methods in Biomedical Engineering, 30(January), 755–766. doi:10.1002/cnm.
Nevalainen, N., Riklund, K., Andersson, M., Axelsson, J., Ögren, M., Lövdén, M., et al. (2015). COBRA: A prospective multimodal imaging study of dopamine, brain structure and function, and cognition. Brain Research, 1612, 83–103. doi:10.1016/j.brainres.2014.09.010.
Nowinski, W. L., Thirunavuukarasuu, A., Volkau, I., Marchenko, Y., Aminah, B., Puspitasari, F., & Runge, V. M. (2009a). A three-dimensional interactive atlas of cerebral arterial variants. Neuroinformatics, 7(4), 255–264. doi:10.1007/s12021-009-9055-0.
Nowinski, W. L., Volkau, I., Marchenko, Y., Thirunavuukarasuu, A., Ng, T. T., & Runge, V. M. (2009b). A 3D model of human cerebrovasculature derived from 3T magnetic resonance angiography. Neuroinformatics, 7(1), 23–36. doi:10.1007/s12021-008-9028-8.
Nowinski, W. L., Chua, B. C., Marchenko, Y., Puspitsari, F., Volkau, I., & Knopp, M. V. (2011). Three-dimensional reference and stereotactic atlas of human cerebrovasculature from 7 Tesla. NeuroImage, 55(3), 986–998. doi:10.1016/j.neuroimage.2010.12.079.
Nowinski, W. L., Puspitasari, F., Volkau, I., Orrison, W. W., & Knopp, M. V. (2013). Quantification of the human cerebrovasculature: a 7Tesla and 320-row CT in vivo study. Journal of Computer Assisted Tomography, 37(1), 117–122.
Osborn, A. G. (1999). Diagnostic cerebral angiography (2nd ed.). Philadelphia: Lippincott Williams & Wilkins.
Palàgyi, K., & Kuba, A. (1998). A 3D 6-subiteration thinning algorithm for extracting medial lines. Pattern Recognition Letters, 19(7), 613–627. doi:10.1016/S0167-8655(98)00031-2.
Passat, N., Ronse, C., Baruthio, J., Armspach, J.-P., Maillot, C., & Jahn, C. (2005). Region-growing segmentation of brain vessels: an atlas-based automatic approach. Journal of Magnetic Resonance Imaging : JMRI, 21(6), 715–725. doi:10.1002/jmri.20307.
Passat, N., Ronse, C., Baruthio, J., Armspach, J. P., & Maillot, C. (2006). Magnetic resonance angiography: from anatomical knowledge modeling to vessel segmentation. Medical Image Analysis, 10(2), 259–274. doi:10.1016/j.media.2005.11.002.
Rivera-Rivera, L. A., Turski, P., Johnson, K. M., Hoffman, C., Berman, S. E., Kilgas, P., et al. (2015). 4D flow MRI for intracranial hemodynamics assessment in Alzheimer’s disease. Journal of Cerebral Blood Flow & Metabolism. doi:10.1177/0271678X15617171.
Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., et al. (2002). Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage, 289, 273–289. doi:10.1006/nimg.2001.0978.
van Baarsen, K. M., Kleinnijenhuis, M., Jbabdi, S., Sotiropoulos, S. N., Grotenhuis, J. A., & van Cappellen van Walsum, A. M. (2016). A probabilistic atlas of the cerebellar white matter. NeuroImage, 124, 724–732. doi:10.1016/j.neuroimage.2015.09.014.
Wåhlin, A., Ambarki, K., Hauksson, J., Birgander, R., Malm, J., & Eklund, A. (2012). Phase contrast MRI quantification of pulsatile volumes of brain arteries, veins, and cerebrospinal fluids compartments: repeatability and physiological interactions. Journal of Magnetic Resonance Imaging, 35(5), 1055–1062. doi:10.1002/jmri.23527.
Wright, S. N., Kochunov, P., Mut, F., Bergamino, M., Brown, K. M., Mazziotta, J. C., et al. (2013). Digital reconstruction and morphometric analysis of human brain arterial vasculature from magnetic resonance angiography. NeuroImage, 82, 170–181. doi:10.1016/j.neuroimage.2013.05.089.
Zarrinkoob, L., Ambarki, K., Wåhlin, A., Birgander, R., Eklund, A., & Malm, J. (2015). Blood flow distribution in cerebral arteries. Journal of Cerebral Blood Flow & Metabolism, 35(4), 648–654. doi:10.1038/jcbfm.2014.241.
Acknowledgements
This study was supported by the Swedish Research Council [grant number 2015–05616], the Swedish Heart and Lung Foundation [grant numbers 20110383, 20140592] and the Swedish Brain Foundation. A grant from the Swedish Research Council [grant number 421-2012-648] to Lars Nyberg supported the COBRA data collection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The local ethical review board approved the COBRA study. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Oral and written information about the study was given and written consent was obtained from all subjects.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dunås, T., Wåhlin, A., Ambarki, K. et al. A Stereotactic Probabilistic Atlas for the Major Cerebral Arteries. Neuroinform 15, 101–110 (2017). https://doi.org/10.1007/s12021-016-9320-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-016-9320-y