Abstract
One of the challenges of modern neuroscience is integrating voluminous data of diferent modalities derived from a variety of specimens. This task requires a common spatial framework that can be provided by brain atlases. The first atlases were limited to two-dimentional presentation of structural data. Recently, attempts at creating 3D atlases have been made to offer navigation within non-standard anatomical planes and improve capability of localization of different types of data within the brain volume. The 3D atlases available so far have been created using frameworks which make it difficult for other researchers to replicate the results. To facilitate reproducible research and data sharing in the field we propose an SVG-based Common Atlas Format (CAF) to store 2D atlas delineations or other compatible data and 3D Brain Atlas Reconstructor (3dBAR), software dedicated to automated reconstruction of three-dimensional brain structures from 2D atlas data. The basic functionality is provided by (1) a set of parsers which translate various atlases from a number of formats into the CAF, and (2) a module generating 3D models from CAF datasets. The whole reconstruction process is reproducible and can easily be configured, tracked and reviewed, which facilitates fixing errors. Manual corrections can be made when automatic reconstruction is not sufficient. The software was designed to simplify interoperability with other neuroinformatics tools by using open file formats. The content can easily be exchanged at any stage of data processing. The framework allows for the addition of new public or proprietary content.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
One of the challenges in the pursuit of understanding brain function is integrating voluminous data of different modalities—histological, functional, electrophysiological, etc.—obtained from different animal models and specimens. To make interpretation of the results accurate or even possible they must be precisely localized in a neuroanatomical context (Bjaalie 2002). Traditionally, this context is provided by 2D brain atlases—collections of graphical representations (drawings and/or photographs) of consecutive brain transsections placed in a spatial coordinate system and providing nomenclature, description and often additional (e.g. neurochemical) characteristics of anatomical structures. Although there are plenty of well established, precise brain atlases they are limited to one or, at the best, three ‘standard’ anatomical planes.
Recent development of modern recording techniques leading to spatially distributed data (magnetic resonance imaging (MRI), positron emission tomography (PET), multichannel local field potential (LFP), gene expression maps, etc.), brought a necessity of three-dimensional brain atlases of various species. Apart from providing a coherent spatial reference for data, 3D brain atlases simplify navigation through brain structures (MacKenzie-Graham et al. 2004), facilitate sectioning at arbitrary angles (Gefen et al. 2005) or designing new cutting planes for in vitro slice preparations containing the desired structures or preserving specific connections. They are also invaluable to position cell models in space, which is needed in modeling measurements of spatially distributed quantities, such as local field potentials (Łęski et al. 2007, 2010; Potworowski et al. 2011).
Three-dimensional atlases have already been constructed from experimental datasets (Neuroterrain—Bertrand and Nissanov (2008), Waxholm Space—Johnson et al. (2010; Hawrylycz (2009; Hawrylycz et al. (2011)), or existing two dimensional reference atlases (e.g. NESYS Atlas3D—Hjornevik et al. (2007), SMART Atlas—Zaslavsky et al. (2004), the Whole Brain Catalog—Larson et al. (2009), CoCoMac-Paxinos3D—Bezgin et al. (2009)). They are usually prepared by manual extraction of the regions of interest from available delineations and creating 3D models in commercial software. Workflows applied in these projects do not allow other researchers to utilize and verify the results easily. It is particularly important in case of the reconstructions made from popular commercial atlases which cannot be freely distributed. So far, no systematic and open approach was offered to enable easy and reproducible creation of 3D models.
Such software should allow for input data of different types and for data exchange with various atlasing systems (e.g. Ruffins et al. 2010; Bakker et al. 2010; Nowinski et al. 2011) and other neuroinformatics projects (e.g. Joshi et al. 2011). The desired features include automation, reproducibility, configurability and transparency. By automatic reconstruction we mean that the user must only provide the input data and specify the parameters of the reconstruction. Errors are logged for further review and do not stop the process which runs without interaction. The user can review results and, depending on the quality of the obtained model and error log, he can correct the input data or change the reconstruction parameters. Full automation is particularly important if on-line applications are considered. Reproducibility means that if the process is repeated with the same input data and parameters it gives identical results. This is in contrast to manual methods: reconstructions for the same input data and parameters done by different people would usually differ. Configurability means that models meeting various requirements can be generated easily. Since both input data and the expected output may vary across different sources and applications, the possibility of extensive process customization is essential. Finally, by transparency of the process we mean the possibility of inspection, analysis and manual correction of the results at any stage.
To address these challenges we present a software package, 3D Brain Atlas Reconstructor (3dBAR), dedicated to automated reconstruction of three-dimensional brain structures from 2D atlases or other compatible data. As a core part of the workflow we introduce a Common Atlas Format (CAF), a general representation of data containing 2D drawings along with additional information needed for transformation into 3D structures, interoperability with other atlasing tools, and other applications. Basic functionality is provided by a set of parsers which translate any 2D data into CAF, and the reconstruction module which extracts structural elements from the CAF dataset and integrates them into a spatial model which can be manipulated in specialized to brain atlases (e.g. NESYS Atlas3D, Slicer3D—Pieper et al. (2006)) or general purpose (we found the Kitware Paraview particularly useful—http://www.paraview.org/) 3D viewers.
To meet the requirements defined above, our workflow is based on free software and open formats (Python environment, Scalable Vector Graphics – SVG, eXtensible Markup Language – XML, Virtual Reality Modeling Language – VRML, Neuroimaging Informatics Technology Initiative – NIfTI format). It can use 2D vector graphics, 2D and 3D raster data, it can also import datasets from other atlasing systems including direct download from the Internet. This procedure is highly automated so reconstructions can be easily repeated, results reviewed, typical errors removed or marked for manual correction. Due to its modular structure our workflow can easily be extended.
Note that the software requires structures’ delineation to be provided as an input. These can be obtained using dedicated tools facilitating automatic segmentation (e.g. Yushkevich et al. 2006; Avants et al. 2011b). The quality of the reconstruction highly depends on the spatial alignment of the input data which can be improved by the registration process done by other specialized software (e.g. Woods et al. 1992; Avants et al. 2011a; Lancaster et al. 2011). Those issues are beyond the scope of presented workflow.
Common Atlas Format
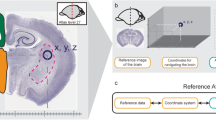
The Common Atlas Format is a format for complete, self-contained storage of processed 2D atlas data. Such data can be used e.g. for generating 3D models or sharing the data with other atlasing systems. The format was designed to maximize interoperability with other software, browsing or incorporating into databases. CAF consists of a set of CAF slides which hold information about shape, names of structures and their locations in a specific (e.g. stereotaxic) spatial coordinate system and of a single index file providing structure hierarchy, holding metadata and summarizing information about all the slides.
The CAF slides are stored as SVG files extended with additional attributes in 3d Brain Atlas Reconstructor XML namespace defined by bar: prefix (see Listing 1 for example). This choice is consistent with the International Neuroinformatics Coordinating Facility (INCF) recommendations for the development of atlasing infrastructure (Hawrylycz 2009, p. 38–39) and has the following advantages: SVG file (even extended with 3dBAR namespace) can be opened by popular graphics software (Inkscape, Adobe Illustrator, Corel Draw, etc.), moreover, a single CAF slide carries delineations and annotations thus no additional data have to be provided to decode file contents. An example of a CAF slide is shown in Fig. 1 with corresponding code in Listing 1.
An example of a CAF slide. The slide was created from the 495th coronal slice of a labeled volume of the Waxholm Space Atlas (Johnson et al. 2010). Original structure colors were preserved. One can distinguish three types of labels: regular labels denoting individual structures; spot labels denoting areas not separated from their parent structures (S1 and Pir – secondary somatosensory and piriform cortices, parts of Cx – cerebral cortex). A comment label with an annotation is placed below the brain outline

Each cut is represented with a single CAF slide. When converting different atlases into CAF if there was a choice in the source of the cutting plane we took coronal slides. The subsequent description of the whole framework follows this assumption although one can take other planes. The spatial location of the slide is stored in transformationmatrix and coronalcoord attributes of bar:data elements. The CAF slide is flat—it contains a single g element with only path and text SVG elements allowed. To simplify exporting, further data processing and to reduce the possibility of errors, coordinates of all the elements must be expressed according to the SVG absolute coordinate system (refer to http://www.w3.org/TR/SVG/paths.html#PathData for details).
Brain structures are represented by SVG closed path elements (as defined by closepath command) filled with color uniquely assigned to the structure and its name encoded in the path id attribute. Note that a given structure may be represented by several paths with common attributes. SVG text elements are used to express three types of labels. Regular labels mark separate regions narrowed by the closed paths (Figs. 1 and 3B). For example, to mark the hippocampus on a slide the label “Hc” is placed within the path delineating this region (Fig. 1). Regular labels and paths are related as each label denotes a particular path. This approach introduces redundancy which allows the cross-validation of the slide and detection of potential inconsistencies. The spot labels which denote only a narrow neighborhood of a spot are used e.g. to mark structures that smoothly go over into others so that it is difficult to draw boundaries between them. This kind of label is also suitable for indicating landmarks. Spot labels begin with a dot. Finally, comment labels, starting with a comma, convey additional information about a region, just like comments on the code in programming languages. They allow adding remarks, information about structure delineation, sharing of comments between people involved in the project, etc., and are ignored in further processing. See Fig. 1 for examples of labels usage.
The index file is an XML document summarizing information about all slides, providing structure hierarchy, and extending the dataset with metadata (Listing 2). Obligatory content of atlasproperties element includes parameters allowing conversion between 2D SVG and spatial coordinate system (RefCoords, ReferenceHeight, ReferenceWidth) and FilenameTemplate for generating filename for particular slide number. It also contains the required set of metadata: timestamp of dataset preparation (CAFCompilationTime), the dataset author’s name and email (CAFCreator, CAFCreatorEmail), name of a given dataset (CAFName), orientation of the slides (CAFSlideOrientation), unit of spatial reference system (CAFSlideUnits) and a general comment field (CAFComment). One can extend atlasproperties with additional metadata such as species of the atlased animal, its sex, age, strain, etc., depending on the needs and availability of that information in a particular source.

slidedetails contains data needed to position each slide in space. The structurelist group contains a summary of the paths extracted from CAF slides: their bounding boxes, unique identifiers (uid) and numbers of slides on which a given structure appears. Hierarchy of structures is stored under a hierarchy element, where each entry (group element) consists of its identifier (id), abbreviation, full name and assigned colour. If an element of the hierarchy has a representation in CAF slides (i.e. there is such a structure among slides), the uid of its representation is attached as another attribute. For example, in Listing 2, in the hierarchy section we see that the cerebral cortex, which is represented directly in CAF slides, has a uid attribute, while the forebrain, which is defined as the sum of other sub-structures has not.
If a different representation of a dataset is required, for instance different structure hierarchies, different color mappings or language versions are needed, one should generate another CAF from the source or from intermediate datasets, such as contour files discussed below, as CAF datasets are not intended to be modified. Note that this is a matter of convention as there is no fundamental difficulty in editing CAF files. However, we feel it is more convenient to work on data on earlier stages. Any change in the CAF slide should be followed by an appropriate update of the index file which may be troublesome when done manually. In our workflow it is handled automatically by parsers creating CAF datasets.
3D Brain Atlas Reconstructor—The Software
3dBAR was developed in Python (http://www.python.org), a powerful, open source, object oriented, cross-platform programming language. These features make it a language of choice in many neuroinformatics projects these days (Davison et al. 2009). XML files were processed using the xml.minidom extension. SVG rasterization and image manipulation was handled by python-rsvg library (http://cairographics.org/pyrsvg/), Python Image Library (PIL – http://www.pythonware.com/products/pil/) and SciPy (http://www.scipy.org/). Graphical User Interface was prepared using WxPython 2.6 (http://www.wxpython.org/).
3D graphics visualization is performed using Visualization ToolKit (VTK, Schroeder et al. (2006), http://www.vtk.org/) which was chosen because it is the best known open-source visualization library, it is well documented, widely used in medical imaging, and implements a wide range of algorithms. VTK is written in C+ +, however it has accessible Python bindings. All segments of the software were prepared in an object-oriented manner to simplify code maintenance and extensibility.
The ultimate goal of the proposed software is semi-automatic generation of 3D models of selected brain structures from their two dimensional representations. The software is divided into three layers where each layer may consist of many interchangeable modules (see Fig. 4).
The first layer, called the Input data layer, consists of components that determine the logical structure of the input data and transform it into the Common Atlas Format given various processing directives and settings provided by the user. As typical datasets are large and have many individual features we found it convenient to encapsulate individual solutions into independent software modules, which we call parsers, one for each dataset considered.
The intermediate layer, called the CAF layer, holds processed input data in the Common Atlas Format. It can be exported and processed in many ways one of which is generating three-dimensional models.
The last layer, called the reconstruction layer, is where 3D models are generated from CAF data using reconstruction parameters such as model resolution, smoothing, output format, etc. The result of this process is a set of three-dimensional models in a form depending on provided settings. We provide graphical and command-line interfaces which simplify processing at this stage (see Section 1 of supplementary materials).
Description of Parsers and their Properties
The first step of the 3dBAR workflow is building a consistent CAF representation of a given data input. To achieve this we have developed several parsers dedicated to specific inputs. Different solutions are used to handle vector and bitmap graphics stored locally, which allows for broad range of processing. Other parsers are used to import and translate preprocessed data from external sources (e.g. other atlasing systems) into CAF.
Vector Processing Workflow
Figure 1 shows a CAF slide (defined in Section “Common Atlas Format”), in which the cross-section of each structure is drawn as a closed SVG path filled with color. Thus, each border is actually defined by two overlaid lines from two paths. To facilitate slide preparation we split processing of vector data into two steps. First we create SVG contour slides (Fig. 3A) in which structures are defined by easily editable contour lines. Once a set of contour slides is available it is transformed into the CAF slides according to the provided processing parameters.
An illustration of the tracing procedure and error correction features implemented in the 3dBAR’s vector workflow. An example contour slide based on Scalable Brain Atlas DB08 dataset, slide 44 (Bakker et al. 2010; Wu et al. 2000) prepared to illustrate the construction of contour slides and the error correction features. (A) A contour slide containing three undefined areas and an open contour (red arrow); spatial coordinate markers are highlighted by red outlines. (B) CAF slide with brain structures represented by closed paths and denoted by labels. Three unlabeled areas were detected and denoted as Unlabeled. The broken contour was closed using the error correction algorithm and the neighboring structures labeled FOG (fronto-orbital gyrus) and LOrG (lateral orbital gyrus) were properly recognized and divided
Contour slides can be created automatically or manually drawn in vector graphics programs (Inkscape, Adobe Ilustrator, Corel Draw). The latter may be necessary, for example, when a new atlas is prepared from scratch or when existing slides require correction. In the case of published atlases, Portable Document Format (PDF) or Postscript files provided by editors may be automatically processed to extract information required to build contour slides. Atlas pages have to be converted to SVG (e.g. using pstoedit, http://www.helga-glunz.homepage.t-online.de/pstoedit/) and then formatted according to the following specification.
A contour slide has to contain a single g element consisting of contours represented by path elements that describe boundaries between structures or other regions of interest. All contours have to be defined with the same color and have to be solid lines but they may vary in thickness. Names of the regions narrowed by contours are indicated by labels. Labels and contours are not related and can be freely modified without concern of data integrity. The contour slide supports the same label types as the CAF slide. Both spot labels and comment labels are disregarded in further processing and copied directly to the CAF slide. Markers are used to localize the slide in a spatial coordinate system. They are text elements with special captions placed at precise locations. In general, one marker is used to determine the position of the slide along a chosen primary axis, in our practice saggital. The other two markers define the dorsal-ventral and lateral-medial coordinates on the coronal plane.
It is convenient to introduce a special label vBrain denoting the complement of the whole brain on every slide. This label has to be placed somewhere outside the actual brain outline. We place it near top-left corner in an area which is unoccupied in every slide. If there are other regions inside the brain outline we wish to exclude (such as closed spaces formed by the cortical folds) additional vBrain labels may be placed during contour slide preparation.
Edition of contour slides gives an opportunity to customize a given atlas to particular needs (e.g. embed experimental results like lesion or staining outlines). To add a new structure one creates a label over a given region bounded by contours. Splitting the structure into smaller substructures comes down to drawing dividing lines or new contours and placing corresponding labels. Redefining the shape of a structure is equivalent to editing its contour.
The correctly prepared contour slide is transformed to a CAF slide by means of a tracing procedure (Fig. 4) which converts contours and labels into closed paths of different colors representing brain structures.
First, spatial information is extracted from markers and expressed using CAF-specific XML elements. All slides are aligned to a common spatial grid so a single set of parameters is needed to transform the coordinates from SVG drawing to the spatial system.
To trace a contour slide we separate labels from contours, which are then rasterized with configurable resolution (note that large image dimensions are required) to a grayscale bitmap and stored in memory. Next, if required, an error correction mechanism is initialized. It consists of an algorithm automatically closing small holes in contours between structures, an algorithm that recognizes labels placed outside brain outline or directly on the contours and detection of unlabeled and duplicate regions. Then, for each stored label, its location (anchor point) is taken, a temporary copy of the rasterized slide is created and a flood-fill algorithm is applied at the point where a respective label is anchored The result of this procedure is an image with gray contours and a black region which is then binarized (if pixel is black, it remains black, otherwise it is changed to white). The binarized image is sent to PoTrace (http://potrace.sourceforge.net/) and traced with customizable parameters. The output is a closed SVG path element representing the region narrowed by the contours. The obtained path is postprocessed and id, name and color attributes are assigned according to the user-provided parameters.
The first labels to be processed are vBrain and the complement of resulting paths gives the outline of the whole brain and is denoted as Brain. This structure is used as a reference to determine unlabeled areas and to detect if a given label points outside the brain outline. Then all the other labels are processed and a new CAF slide is created. It consists of traced paths and labels taken from the contour slide and, optionally, new labels indicating unlabeled structures.
The tracing procedure is consecutively applied to all contour slides. Once this is done, the index file is generated. First, for every structure its name and the numbers of slides on which it appears are stored and a bounding box is calculated. Precalculating bounding boxes reduces the amount of time and memory while generating a 3D model. If a hierarchy of structures is provided, it is used to create an ontology tree. Otherwise a flat hierarchy is created—all structures are gathered under superior Brain structure. In addition, one can specify the full name and color for each hierarchy element.
An example of vector processing workflow including contour slide preparation and detailed description of error correcting features can be found in supplementary materials online.
Bitmap Processing Workflow
The input data may come in bitmap form e.g. from segmented magnetic resonance, imaging scans, histology plates where structures are colored rather than defined by their boundaries, or from volumetric datasets (i.e. Nifti or Analyze files). To process such data into CAF we developed another parser. We convert bitmaps into CAF slides because SVG files can hold arbitrary additional information apart from structure delineation and can be easy translated to other types of data such as database entries. Usually, SVG drawings are also smaller in terms of size than their bitmap equivalents.
In stacked bitmaps or volumetric datasets brain structures are encoded as regions with the same unique value. To decode these values into specific colors a look-up table is required. It is often provided as part of the source data. Otherwise we use an additional tab-separated text file which holds the table assigning structure labels and colors to volume indices. Optionally, as in a vector parser, one can provide additional data, such as a hierarchy and full structure names. Finally, the voxel dimensions and the origin of the spatial coordinate system must be provided. Such information is usually available in volumetric datasets, but in case of stacked bitmaps, there is no internal spatial reference and it has to be defined by the user. The parser assumes that all the slides are aligned, that is they have the same spatial coordinates of corners in the coronal plane. However, they do not have to be uniformly distributed along the anterior-posterior axis.
Bitmaps are processed directly into CAF slides without the intermediate contour slide stage. The parsing procedure starts with loading the necessary input data consisting of color codes and spatial reference of the source dataset. Then stacked bitmaps or slices extracted from the volumetric dataset are processed one by one. All colors present in the analyzed bitmap are identified and a binary mask is created for each of them. By default, patches smaller than a given, customizable, number of pixels are skipped. Each mask is sent to PoTrace where it undergoes the tracing procedure. Resulting SVG path is then post-processed by setting its attributes such as id, name of corresponding structure, color, etc., and a regular label related to the path is created. Final CAF slides undergo indexing routine (see vector processing workflow) which completes generation of a CAF dataset.
Exchanging Content with External Atlasing Systems
With the development of digital atlasing infrastructure more and more often one wants to interact with external tools and process data available remotely. To achieve this goal we have implemented mechanisms facilitating import and export of data from/to external tools, databases or web pages. Data may be exchanged on the level of CAF dataset or of final 3D reconstructions saved in the form of volumetric dataset or polygonal mesh. As an example we have prepared a parser which allows data exchange between 3dBAR and ScalableBrainAtlas (SBA, Bakker et al. (2010), http://scalablebrainatlas.incf.org, Fig. 12) atlasing systems which both use SVG-based storage format. The parser always downloads the most recent version of the chosen SBA template and then converts it into CAF dataset. There is no need for additional input since SBA templates include all the necessary information.
The interaction with SBA is bidirectional as CAF data from 3dBAR may be exported and displayed in ScalableBrainAtlas although CAF does not contain complete information required for full functionality of SBA services. The interoperability between 3dBAR and other neuroinformatics projects will be further explored.
Reconstruction
The main purpose of 3d Brain Atlas Reconstructor is building 3D models of brain regions. This is facilitated by graphical and command-line based tools (see supplementary materials). The process of reconstruction relies on successive filling a bounding box with the structure of interest and locating this volume in a spatial coordinate system. Detailed reconstruction workflow is presented in Fig. 5.
An execution of reconstruction routine creates a model of a single hierarchy element. If it contains substructures, they will all be merged (Fig. 6A, B). Technically, a set of all the identifiers of the selected structure and its substructures is created and a list of all the slides containing at least one of them is built. Dimensions of a single voxel are defined separately in the coronal plane and along the anterior-posterior axis. The size of the bounding box to be filled with the reconstructed model depends on a given resolution which also controls reconstruction accuracy and level of detail to be achieved. To use all the available data the anterior-posterior resolution should not exceed the distance between two consecutive slides. For smaller resolution some slides may be skipped. On the other hand, selecting higher resolution results in more detailed reconstructions but longer processing times. Thus it is recommended to adapt the resolution individually for each dataset and selected structures.
An exemplary reconstruction of cerebral cortex structure consisting of various cortical areas (Macaque monkey, CAF dataset created using Scalable Brain Atlas DB08 template, Wu et al. (2000)). (A) An exemplary CAF slide containing cortical fields and other structures not belonging to cerebral cortex. (B) Cortical areas available on the given slide according to a provided hierarchy are extracted. (C) Cerebral cortex binary mask is created by merging the cortical areas. (D) The depth is assigned to all the masks (it may vary across slides) which are then stacked into a volume. Note that spaces between consecutive masks were added for figure clarity while in the actual reconstruction the masks fill continuously entire bounding box (only the first 25 of 161 masks are presented). (E) A volumetric rendering of reconstructed hemisphere of cerebral cortex. A part of the model was cut off to emphasize the volumetric nature of the reconstruction
A set of bitmap masks is created by rasterizing consecutive slides (Fig. 6C). The height and the width of these masks is based on the maximal extent of the structure in coronal planes. A depth of each mask (span along the anterior-posterior dimension) may vary since it is defined by the distance between consecutive slides (Fig. 6D). The entire bounding box is filled with masks creating volumetric representation of the given structure (Fig. 6E).
The volume prepared this way is further shaped with the support of VTK visualization library pipeline (Fig. 7). After optional anisotropic Gaussian smoothing of input volume (vtkImageGaussianSmooth) the surface of the structure is extracted using a marching cubes algorithm with a given threshold value (vtkMarchingCubes). The extracted polygonal mesh may then be smoothed using Laplacian smoothing (vtkSmoothPolyDataFilter). If necessary, it can be compressed using vtkQuadricClustering filter. This allows us to eliminate unnecessary polygons and vertices reducing model complexity and size of the output file. Additionally, if the CAF dataset has defined only one hemisphere, it is possible to mirror it to create the second hemisphere (vtkTransformPolyDataFilter). The final reconstructed model can be exported as a volumetric dataset or polygonal mesh.
If we define reconstruction error as the maximum distance between corresponding points of the contour in the CAF slide and in the reconstruction created using default reconstruction settings (isosurface extraction using the threshold value of 128 in the marching cubes algorithm and no further mesh processing) it will always be smaller than twice the voxel resolution along a given axis.
Results
During development and testing of 3d Brain Atlas Reconstructor we have prepared several CAF datasets based on three types of source atlas: a PDF file, a volumetric dataset and on data derived from external atlasing systems.
The biggest challenge was to derive two datasets from PDF files containing digital editions of published printed atlases, The Rat Brain in Stereotaxic Coordinates, 6th edition (Paxinos and Watson 2007) and The Mouse Brain in Stereotaxic Coordinates (Paxinos and Franklin 2008). Slides in those atlases consist of contours delineating the whole brain into separated regions and their labels which makes them suitable for vector workflow. While similar in general format these atlases differ significantly in details. Because of this each source atlas was processed using a separate parser derived from the generic vector parser described in Section “Vector Processing Workflow”. Consecutive stages of processing and sample reconstructions are presented in Fig. 8. The results are in good comparison with analogous reconstructions created by Hjornevik et al. (2007) using different workflow (Fig. 9). The mouse data required significant manual corrections to achieve satisfactory models. Comparison of reconstruction before and after this process is shown in Fig. 10. CAF datasets from both atlases can still be refined by further users.
Various examples of rat brain reconstructions. (A) A contour slide created by parsing the source atlas reproduced from Paxinos and Watson (2007), with permission. (B) An example of a CAF slide containing 62 structures. Five paths and five labels are denoted as Unlabelled and gathered as the Unlabelled structure. (C) The thalamus decomposed up to the first level and the pyramidal tract. Both models presented as polygonal meshes. (D) A horizontal cut of volumetric representation of the thalamus. Each colour represents a different substructure. (E) The hippocampal formation presented in the form of a polygonal mesh; additional smoothing was applied. (F) An analogous reconstruction without any additional mesh processing. The reconstruction is decomposed up to the first level of substructures. The pale green model hippocampus, brown enthorinal cortex, green subiculum, gray postsubiculum, olive parasubiculum
Reconstructions of the mouse brain structures based on Paxinos and Franklin (2008) showing distortions caused by data inconsistencies, particularly by leaking structures. (A) A comparison of two reconstructions of amygdala: left before, right after manual corrections to the contour slides. The number of deformations is significantly reduced and the shape of the reconstruction is much closer to expectations. (B) Two models of Lateral septal nucleus left before manual corrections, shows a minor deformation of the model, right the same structure after manual corrections. (C) The isocortex reconstructed as the parent structure with the first level of substructures according to a given ontology tree
These atlases come with supplementary data such as full names of structures, however, both of them lack ontology trees binding all structures into consistent hierarchies. As we did not find ontologies completely adapted to any of those two atlases we created a hierarchy covering the majority of structures by combining databases from the NeuroNames ((Bowden and Dubach 2003), an ontology for Macaca fascicularis), Brain Architecture Management System (BAMS, Bota et al. (2005), ontology trees based on various atlases), and by including our neuroanatomical knowledge.
The second group of CAF atlases was derived from volumetric data. One dataset was obtained from the atlas of C57BL/6 mouse brain (Johnson et al. 2010) based on MRI and Nissl histology introducing the Waxholm Space—the proposed reference coordinate system for the mouse brain. The volume containing 37 structures used for creating the CAF dataset is available at the INCF Software Center (http://software.incf.org/software/waxholm-space) and was extended with a simple hierarchy. Reconstructions created by 3dBAR using this template are also localized in the Waxholm Space spatial reference system.
Another dataset derived from a volumetric source atlas is the average-shape atlas of the honeybee brain (Brandt et al. 2005) created using confocal imaging of 20 specimens of honeybee brains and delineated using average-shape algorithm. The data used for creation of the CAF dataset were downloaded from http://www.neurobiologie.fu-berlin.de/beebrain. Examples of reconstructions based on these volumetric datasets are shown in Fig. 11.
The last group of available CAF datasets were created using interoperability with the Scalable Brain Atlas. Three Scalable Brain Atlas templates were converted into CAF: Paxinos Rhesus Monkey atlas (PHT00, Paxinos et al. (2000)), NeuroMaps Macaque atlas (DB08, Wu et al. (2000)) and Waxholm Space for the mouse (WHS09, Johnson et al. (2010)). Note that WHS09 dataset is different from the volumetric dataset defining the Waxholm space in form and in content being derived from a sample of the original. Figure 12 shows exemplary reconstructions from SBA datasets.
Reconstructions based on CAF datasets derived from Scalable Brain Atlas templates (Bakker et al. 2010). (A–C) A macaque brain created using DB08 (Wu et al. 2000) template. (D–E) Rhesus brain based on PHT00 (Paxinos et al. 2000) template. (A) Separated cortical gyri in the form of a polygonal mesh (B) A volumetric representation of full-depth model of neocortex. Each defined substructure is represented using a different color. Part of the left hemisphere was cut off to visualize a cross section of the reconstruction. (C) The thalamus in the form of a polygonal mesh without additional processing. (D) The parcelated isocortex, (E) Exemplary subcortical structures: the thalamus, the amygdala and the basal ganglia
Summary and Outlook
We have designed and implemented a workflow dedicated to processing two dimensional data of different quality, complexity and origin, into three dimensional reconstructions of brain structures. We have also proposed a common format for these data, convenient for our purposes but of broader applicability, which we called the Common Atlas Format (CAF). Every dataset in CAF consists of an XML index file and SVG slides representing brain slices. Each slide contains a decomposition of a brain slice into separate structures represented by closed paths and holds information about the spatial coordinate system in which the brain is located. The CAF index file has an embedded ontology tree binding all the structures into a consistent hierarchy and may include additional information such as full names of structures or external references to databases. All presented parsers and CAF dataset elements (slide, structures, labels, etc.) were implemented in a Python module with an application programming interface (API) allowing manipulation of CAF datasets and creation of new parsers by extending generic classes.
The reconstruction process leading to a three dimensional model operates in two steps. The first step is parsing the source atlas and producing a CAF dataset. Any data containing consistent information about brain structures analogous to CAF content may be processed using one of the provided parsers or by deriving one for a new format. The second stage involves processing CAF datasets and results in a complete 3D reconstruction in the form of a volumetric dataset or polygonal mesh obtained with the support of VTK Visualization Toolkit.
The presented workflow leads to highly-automated and reproducible reconstructions, and it can be customized as needed. Moreover, it enables tracking and reviewing of the whole reconstruction process as well as locating and eliminating potential reconstruction errors or data inconsistencies. We have used this workflow to process seven source atlases. Two of them were based on PDF files containing digital versions of published printed atlases. Another two were prepared from volumetric datasets and the other three derived from Scalable Brain Atlas (Bakker et al. 2010). The proposed workflow can be extended to accomodate source data in additional formats requiring only a new parser for each format. The reconstruction process was wrapped with a GUI resulting in a fully functional application which can be used for loading CAF datasets, generating and exporting reconstructions and allowing fine-tuning of the reconstruction process.
Clearly, in order to produce reasonable reconstructions, 3dBAR needs input data of good quality. This requires careful processing of raw data including precise segmentation and alignment. However, since these issues are addressed by other dedicated, open software (e.g. Woods et al. 1992, Avants et al. 2011a, Lancaster et al. 2011) we skip them in our workflow.
In further development of the software, we consider implementing more sophisticated slice interpolation algorithms (i.e. Barrett et al. 1994; Cohen-or and Levin 1996; Braude et al. 2007) as the naive algorithm currently implemented only assigns thickness to the slices without any interpolation in between. We develop an optimized version of 3d Brain Atlas Reconstructor as an on-line service (available at http://service.3dbar.org/). It provides a browser-based interface with reconstruction module and access to hosted datasets and models. It also accepts direct HTTP queries which simplifies interaction with external software. We intend to integrate this service with the INCF digital atlasing infrastructure.
Another challenge is the distribution of reconstructed structures or CAF datasets. It requires solving practical and—for some data—legal issues. Our software has been designed to be data-agnostic as much as possible. As a result, any owner of compatible data may generate a CAF and models of structures of interest and decide what and how to share. This applies also to commercially available datasets (see Information Sharing Statement, below).
Information Sharing Statement
3d Brain Atlas Reconstructor software with a selection of parsers and a repository of reconstructions is available through the INCF Software Center and the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC). Visit http://www.3dbar.org for release announcements. Supplementary materials containing the description of the GUI as well as the discussion of error correction in the vector parser are available at http://www.3dbar.org/wiki/barSupplement.
The external software we used and the source datasets are available at the locations given within the text with the exception of CAF datasets created from Paxinos and Watson (2007) and Paxinos and Franklin (2008) which are based on proprietary data and have restricted copyrights. Users owning legal copies of these atlases can prepare CAF datasets and reconstructions by themselves using dedicated parsers and hierarchies provided with the 3d Brain Atlas Reconstructor distribution. The authors may be contacted for details.
References
Avants, B. B., Tustison, N. J., Song, G., Cook, P. A., Klein, A., & Gee, J. C. (2011a). A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage, 54(3), 2033–2044.
Avants, B. B., Tustison, N. J., Wu, J., Cook, P. A., & Gee, J. C. (2011b). An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics, 9(4), 381–400. doi:10.1007/s12021-011-9109-y.
Bakker, R., Larson, S. D., Strobelt, S., Hess, A., Wójcik, D. K., Majka, P., & Kötter, R. (2010). Scalable brain atlas: From stereotaxic coordinate to delineated brain region. Frontiers in Neuroscience, 4, 5.
Barrett, W., Mortensen, E., & Taylor, D. (1994). An image space algorithm for morphological contour interpolation. In Proc. graphics interface.
Bertrand, L., & Nissanov, J. (2008). The neuroterrain 3d mouse brain atlas. Frontiers in Neuroinformatics, 2(7), 3.
Bezgin, G., Reid, A., Schubert, D., & Kötter, R. (2009). Matching spatial with ontological brain regions using Java tools for visualization, database access, and integrated data analysis. Neuroinformatics, 7, 7–22. doi:10.1007/s12021-008-9039-5.
Bjaalie, J. G. (2002). Opinion: Localization in the brain: New solutions emerging. Nature Reviews, Neuroscience, 3(4), 322–325.
Bota, M., Dong, H.-W., & Swanson, L. (2005). Brain architecture management system. Neuroinformatics, 3, 15–47.
Bowden, D., & Dubach, M. (2003). Neuronames 2002. Neuroinformatics, 1, 43–59.
Brandt, R., Rohlfing, T., Rybak, J., Krofczik, S., Maye, A., Westerhoff, M., et al. (2005). Three-dimensional average-shape atlas of the honeybee brain and its applications. Journal of Comparative Neurology, 492(1), 1–19.
Braude, I., Marker, J., Museth, K., Nissanov, J., & Breen, D. (2007). Contour-based surface reconstruction using mpu implicit models. Graphical Models, 69(2), 139–157.
Cohen-or, D., & Levin, D. (1996). Guided multi-dimensional reconstruction from cross-sections. In Advanced topics in multivariate approximation.
Davison, A. P., Hines, M. L., & Muller, E. (2009). Trends in programming languages for neuroscience simulations. Frontiers in Neuroscience, 3(3), 374–380.
Gefen, S., Bertrand, L., Kiryati, N., & Nissanov, J. (2005). Localization of sections within the brain via 2d to 3d image registration. In Proceedings ICASSP 05 IEEE international conference on acoustics speech and signal processing 2005 (pp. 733–736).
Hawrylycz, M. (2009). The incf digital atlasing program: Report on digital atlasing standards in the rodent brain. Nature Precedings. doi:10.1038/npre.2009.4000.1
Hawrylycz, M., Baldock, R. A., Burger, A., Hashikawa, T., Johnson, G. A., Martone, M., et al. (2011). Digital atlasing and standardization in the mouse brain. PLoS Computational Biology, 7(2), e1001065. doi:10.1371/journal.pcbi.1001065
Hjornevik, T., Leergaard, T. B., Darine, D., Moldestad, O., Dale, A. M., Willoch, F., et al. (2007). Three-dimensional atlas system for mouse and rat brain imaging data. Frontiers in Neuroinformatics, 1, 4.
Johnson, G. A., Badea, A., Brandenburg, J., Cofer, G., Fubara, B., Liu, S., et al. (2010). Waxholm space: An image-based reference for coordinating mouse brain research. NeuroImage, 53(2), 365–72.
Joshi, A., Scheinost, D., Okuda, H., Belhachemi, D., Murphy, I., Staib, L. H., et al. (2011). Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics, 9(1), 69–84.
Lancaster, J., McKay, D., Cykowski, M., Martinez, M., Tan, X., Valaparla, S., et al. (2011). Automated analysis of fundamental features of brain structures. Neuroinformatics. doi:10.1007/s12021-011-9108-z
Larson, S., Aprea, C., Maynard, S., Peltier, S., Martone, M., & Ellisman, M. (2009). The whole brain catalog. In Neuroscience meeting planner. Chicago, IL: Society for neuroscience, Program No. 895.26.
Łęski, S., Kublik, E., Świejkowski, D., Wróbel, A., & Wójcik, D. K. (2010). Extracting functional components of neural dynamics with ica and icsd. Journal of Computational Neuroscience, 29, 459–473.
Łęski, S., Wójcik, D. K., Tereszczuk, J., Świejkowski, D., Kublik, E., & Wróbel, A. (2007). Inverse current-source density in three dimensions. Neuroinformatics, 5, 207–222.
MacKenzie-Graham, A., Lee, E.-F., Dinov, I. D., Bota, M., Shattuck, D. W., Ruffins, S., et al. (2004). A multimodal, multidimensional atlas of the c57bl/6j mouse brain. Journal of Anatomy, 204(2), 93–102.
Nowinski, W., Chua, B., Yang, G., & Qian, G. (2011). Three-dimensional interactive and stereotactic human brain atlas of white matter tracts. Neuroinformatics. doi:10.1007/s12021-011-9118-x
Paxinos, G. & Franklin, K. B. J. (2008). The mouse brain in stereotaxic coordinates. Elsevier, 3rd Edn.
Paxinos, G., Huang, X.-F., & Toga, A. W. (2000). The Rhesus Monkey brain in stereotaxic coordinates. Academic Press.
Paxinos, G., & Watson, C. (2007). The rat brain in stereotaxic coordinates. Elsevier, 6th Edn.
Pieper, S., Lorensen, B., Schroeder, W., & Kikinis, R. (2006). The na-mic kit: Itk, vtk, pipelines, grids and 3d slicer as an open platform for the medical image computing community. In Proc. IEEE intl. symp. on biomedical imaging ISBI: From nano to macro (pp. 698–701). IEEE.
Potworowski, J., Głąbska, H., Łęski, S., and Wójcik, D. (2011). Extracting activity of individual cell populations from multielectrode recordings. BMC Neuroscience, 12(Suppl 1), P374.
Ruffins, S. W., Lee, D., Larson, S. D., Zaslavsky, I., Ng, L., & Toga, A. (2010). Mbat at the confluence of Waxholm space. Frontiers in Neuroscience, 4(0), 5.
Schroeder, W., Martin, K., & Lorensen, B. (2006). Visualization toolkit: An object-oriented approach to 3D graphics, 4th Edn. Kitware.
Woods, R. P., Cherry, S. R., & Mazziotta, J. C. (1992). Rapid automated algorithm for aligning and reslicing pet images. Journal of Computer Assisted Tomography, 16, 620–633.
Wu, J., Dubach, M., Robertson, J., Bowden, D., & Martin, R. (2000). Primate brain maps: Structure of the Macaque brain: A laboratory guide with original brain sections. Printed Atlas and Electronic Templates for Data and Schematics, Elsevier Science, 1st Edn.
Yushkevich, P. A., Piven, J., Hazlett, H. C., Smith, R. G., Ho, S., Gee, J. C., et al. (2006). User-guided 3d active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage, 31(3), 1116–1128.
Zaslavsky, I., He, H., Tran, J., Martone, M. E., & Gupta, A. (2004). Integrating brain data spatially: Spatial data infrastructure and atlas environment for online federation and analysis of brain images. In DEXA workshops (pp. 389–393).
Acknowledgements
We are grateful to Jan Bjaalie and Rembrandt Bakker for illuminating discussions during various stages of this project. In particular, different types of labels arose in discussions of PM with RB. We are grateful to Mark Hunt and Greg McNevin for their comments on the manuscript. This work was supported by a project co-financed by the European Regional Development Fund under the Operational Programme Innovative Economy, POIG.02.03.00-00-003/09, and by a travel grant from International Neuroinformatics Coordinating Facility to PM.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Majka, P., Kublik, E., Furga, G. et al. Common Atlas Format and 3D Brain Atlas Reconstructor: Infrastructure for Constructing 3D Brain Atlases. Neuroinform 10, 181–197 (2012). https://doi.org/10.1007/s12021-011-9138-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-011-9138-6