Abstract
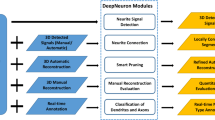
Automating the process of neural circuit reconstruction on a large-scale is one of the foremost challenges in the field of neuroscience. In this study we examine the methodology for circuit reconstruction from three-dimensional light microscopy (LM) stacks of images. We show how the minimal error-rate of an ideal reconstruction procedure depends on the density of labeled neurites, giving rise to the fundamental limitation of an LM based approach for neural circuit research. Circuit reconstruction procedures typically involve steps related to neuron labeling and imaging, and subsequent image pre-processing and tracing of neurites. In this study, we focus on the last step—detection of traces of neurites from already pre-processed stacks of images. Our automated tracing algorithm, implemented as part of the Neural Circuit Tracer software package, consists of the following main steps. First, image stack is filtered to enhance labeled neurites. Second, centerline of the neurites is detected and optimized. Finally, individual branches of the optimal trace are merged into trees based on a cost minimization approach. The cost function accounts for branch orientations, distances between their end-points, curvature of the merged structure, and its intensity. The algorithm is capable of connecting branches which appear broken due to imperfect labeling and can resolve situations where branches appear to be fused due the limited resolution of light microscopy. The Neural Circuit Tracer software is designed to automatically incorporate ImageJ plug-ins and functions written in MatLab and provides roughly a 10-fold increases in speed in comparison to manual tracing.









Similar content being viewed by others
References
Al-Kofahi, K. A., Lasek, S., Szarowski, D. H., Pace, C. J., Nagy, G., Turner, J. N., et al. (2002). Rapid automated three-dimensional tracing of neurons from confocal image stacks. IEEE Transactions on Information Technology in Biomedicine, 6, 171–187.
Bas E, Erdogmus D. (2010a). Piecewise linear cylinder models for 3-dimensional axon segmentation in Brainbow imagery. Pages 1297–1300. Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on.
Bas, E., & Erdogmus, D. (2010b). Principal curve tracing. Pages 405–410. ESANN 2010 proceedings, European Symposium on Artificial Neural Networks—Computational Intelligence and Machine Learning. Bruges, Belgium.
Bohland, J. W., et al. (2009). A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Computational Biology, 5, e1000334.
Boyd, S. P., & Vandenberghe, L. (2004). Convex optimization. Cambridge: Cambridge University Press.
Braitenberg, V., & Schüz, A. (1998). Cortex: statistics and geometry of neuronal connectivity. Berlin: Springer.
Briggman, K. L., & Denk, W. (2006). Towards neural circuit reconstruction with volume electron microscopy techniques. Current Opinion in Neurobiology, 16, 562–570.
Brown, K. M., Barrionuevo, G., Canty, A. J., De Paola, V., Hirsch, J. A., Jefferis, G. S., et al. (2011). The DIADEM Data Sets: Representative Light Microscopy Images of Neuronal Morphology to Advance Automation of Digital Reconstructions. Neuroinformatics.
Can, A., Shen, H., Turner, J. N., Tanenbaum, H. L., & Roysam, B. (1999). Rapid automated tracing and feature extraction from retinal fundus images using direct exploratory algorithms. IEEE Transactions on Information Technology in Biomedicine, 3, 125–138.
Cannon, R. C., Turner, D. A., Pyapali, G. K., & Wheal, H. V. (1998). An on-line archive of reconstructed hippocampal neurons. Journal of Neuroscience Methods, 84, 49–54.
Chen, B. L., Hall, D. H., & Chklovskii, D. B. (2006). Wiring optimization can relate neuronal structure and function. Proceedings of the National Academy of Sciences of the United States of America, 103, 4723–4728.
De Paola, V., Holtmaat, A., Knott, G., Song, S., Wilbrecht, L., Caroni, P., et al. (2006). Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron, 49, 861–875.
Denk, W., & Horstmann, H. (2004). Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biology, 2, e329.
Deschamps, T., & Cohen, L. D. (2001). Fast extraction of minimal paths in 3D images and applications to virtual endoscopy. Medical Image Analysis, 5, 281–299.
Dijkstra, E. W. (1959). A note on two problems in connexion with graphs. Numerische Mathematik, 1, 269–271.
Engel, A., & Broeck, Cvd. (2001). Statistical mechanics of learning. Cambridge: Cambridge University Press.
Escobar, G., Fares, T., & Stepanyants, A. (2008). Structural plasticity of circuits in cortical neuropil. The Journal of Neuroscience, 28, 8477–8488.
Frangi, A. F., Niessen, W. J., Vincken, K. L., Viergever, M. A. (1998). Multiscale vessel enhancement filtering. Medical Image Computing and Computer-Assisted Intervention - Miccai'98 1496: pp. 130–137.
Freeman, W. T., Adelson, E. H., & Massachusetts Institute of Technology. Media Laboratory. Vision and Modeling Group. (1991). The design and use of steerable filters. Cambridge: Vision and Modeling Group, Media Laboratory, Massachusetts Institute of Technology.
Gillette, T. A., Brown, K. M., & Ascoli, G. A. (2011). The DIADEM metric: comparing multiple reconstructions of the same neuron. Neuroinformatics.
Gonzalez, G., Aguet, F., Fleuret, F., Unser, M., & Fua, P. (2009). Steerable features for statistical 3D dendrite detection. Med Image Comput Comput Assist Interv, 12, 625–632.
González, G., Fleuret, F., & Fua, P. (2008). Automated delineation of dendritic networks in noisy image stacks. In D. Forsyth, P. Torr, & A. Zisserman (Eds.), Computer vision—ECCV 2008 (Vol. 5305, pp. 214–227). Berlin: Springer.
González G, Turetken E, Fleuret F, Fua P. (2010). Delineating trees in noisy 2D images and 3D image-stacks. Pages 2799–2806. Computer Vision and Pattern Recognition (CVPR), 2010 IEEE Conference on.
Grutzendler, J., Kasthuri, N., & Gan, W. B. (2002). Long-term dendritic spine stability in the adult cortex. Nature, 420, 812–816.
Hayworth, K. J., Kasthuri, N., Schalek, R., & Lichtman, J. W. (2006). Automating the collection of ultrathin serial sections for large volume TEM reconstructions. Microscopy and Microanalysis, 12, 86–87.
Holtmaat, A. J., Trachtenberg, J. T., Wilbrecht, L., Shepherd, G. M., Zhang, X., Knott, G. W., et al. (2005). Transient and persistent dendritic spines in the neocortex in vivo. Neuron, 45, 279–291.
Jacob, M., & Unser, M. (2004). Design of steerable filters for feature detection using canny-like criteria. IEEE Transactions on Pattern Analysis and Machine Intelligence, 26, 1007–1019.
Jefferis, G. S., Potter, C. J., Chan, A. M., Marin, E. C., Rohlfing, T., Maurer, C. R., Jr., et al. (2007). Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell, 128, 1187–1203.
Kass, M., Witkin, A., & Terzopoulos, D. (1988). Snakes: active contour models. International Journal of Computer Vision, 1, 321–331.
Lee, T. C., Kashyap, R. L., & Chu, C. N. (1994). Building skeleton models via (3-D) medial surface/axis thinning algorithms. CVGIP: Graph. Models and Image Processing, 56, 462–478.
Lichtman, J. W., & Sanes, J. R. (2008). Ome sweet ome: what can the genome tell us about the connectome? Current Opinion in Neurobiology, 18, 346–353.
Lichtman, J. W., Livet, J., & Sanes, J. R. (2008). A technicolour approach to the connectome. Nature Reviews. Neuroscience, 9, 417–422.
Livet, J., Weissman, T. A., Kang, H., Draft, R. W., Lu, J., Bennis, R. A., et al. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature, 450, 56–62.
Lorenz, C., Carlsen, I. C., Buzug, T. M., Fassnacht, C., & Weese, J. (1997). Multi-scale line segmentation with automatic estimation of width, contrast and tangential direction in 2D and 3D medical images. Cvrmed-Mrcas’97. 1205: pp. 233–242.
Lu, J., Tapia, J. C., White, O. L., & Lichtman, J. W. (2009). The interscutularis muscle connectome. PLoS Biology, 7, e32.
Marr, D., & Hildreth, E. (1980). Theory of edge detection. Proceedings of the Royal Society of London. Series B: Biological Sciences, 207, 187–217.
Meijering, E. (2010). Neuron tracing in perspective. Cytometry. Part A, 77, 693–704.
Mishchenko, Y., Hu, T., Spacek, J., Mendenhall, J., Harris, K. M., & Chklovskii, D. B. (2010). Ultrastructural analysis of hippocampal neuropil from the connectomics perspective. Neuron, 67, 1009–1020.
Palagyi, K., & Kuba, A. (1998). A 3D 6-subiteration thinning algorithm for extracting medial lines. Pattern Recognition Letters, 19, 613–627.
Russ, J. C. (2007). The image processing handbook. Boca Raton: CRC/Taylor and Francis.
Sato, Y., Nakajima, S., Shiraga, N., Atsumi, H., Yoshida, S., Koller, T., et al. (1998). Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Medical Image Analysis, 2, 143–168.
Sethian, J. A. (1999). Level set methods and fast marching methods: evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science. Cambridge: Cambridge University Press.
Sporns, O., Tononi, G., & Kotter, R. (2005). The human connectome: a structural description of the human brain. PLoS Computational Biology, 1, e42.
Srinivasan, R., Zhou, X., Miller, E., Lu, J., Litchman, J., & Wong, S. T. (2007). Automated axon tracking of 3D confocal laser scanning microscopy images using guided probabilistic region merging. Neuroinformatics, 5, 189–203.
Stepanyants, A., & Chklovskii, D. B. (2005). Neurogeometry and potential synaptic connectivity. Trends in Neurosciences, 28, 387–394.
Stepanyants, A., Hof, P. R., & Chklovskii, D. B. (2002). Geometry and structural plasticity of synaptic connectivity. Neuron, 34, 275–288.
Stepanyants, A., Tamas, G., & Chklovskii, D. B. (2004). Class-specific features of neuronal wiring. Neuron, 43, 251–259.
Stepanyants, A., Martinez, L. M., Ferecsko, A. S., & Kisvarday, Z. F. (2009). The fractions of short- and long-range connections in the visual cortex. Proceedings of the National Academy of Sciences of the United States of America, 106, 3555–3560.
Stepanyants, A., Hirsch, J. A., Martinez, L. M., Kisvarday, Z. F., Ferecsko, A. S., & Chklovskii, D. B. (2008). Local potential connectivity in cat primary visual cortex. Cerebral Cortex, 18, 13–28.
Streekstra, G. J., & van Pelt, J. (2002). Analysis of tubular structures in three-dimensional confocal images. Network, 13, 381–395.
Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E., et al. (2002). Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature, 420, 788–794.
Vasilkoski, Z., & Stepanyants, A. (2009). Detection of the optimal neuron traces in confocal microscopy images. Journal of Neuroscience Methods, 178, 197–204.
Wang, J., Zhou, X., Lu, J., Lichtman, J., Chang, S.-F., Wong, S. T. C. (2007). Dynamic local tracing for 3D axon curvilinear structure detection from microscopic image stack. IEEE, International Symposium of Biomedical Imaging (ISBI). Washington D. C.
Weaver, C. M., Hof, P. R., Wearne, S. L., & Lindquist, W. B. (2004). Automated algorithms for multiscale morphometry of neuronal dendrites. Neural Computation, 16, 1353–1383.
White, J., Southgate, E., Thomson, J., & Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 314, 1–340.
Wilt, B. A., Burns, L. D., Wei Ho, E. T., Ghosh, K. K., Mukamel, E. A., & Schnitzer, M. J. (2009). Advances in light microscopy for neuroscience. Annual Review of Neuroscience, 32, 435–506.
Wink, O., Frangi, A. F., Verdonck, B., Viergever, M. A., & Niessen, W. J. (2002). 3D MRA coronary axis determination using a minimum cost path approach. Magnetic Resonance in Medicine, 47, 1169–1175.
Xie, J., Zhao, T., Lee, T., Myers, E., & Peng, H. (2010). Automatic neuron tracing in volumetric microscopy images with anisotropic path searching. Med Image Comput Comput Assist Interv, 13, 472–479.
Yuste, R., & Bonhoeffer, T. (2001). Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annual Review of Neuroscience, 24, 1071–1089.
Zhou, Y., & Toga, A. W. (1999). Efficient skeletonization of volumetric objects. IEEE Transactions on Visualization and Computer Graphics, 05, 196–209.
Zhou, Y., Kaufman, A., & Toga, A. W. (1998). Three-dimensional skeleton and centerline generation based on an approximate minimum distance field. Visual Computer, 14, 303–314.
Acknowledgements
We would like to acknowledge Dr. Zlatko Vasilkoski’s contribution to the implementation of the voxel coding algorithm and discussions related to the subject of this study. This work was supported by the NIH grant NS063494.
Author information
Authors and Affiliations
Corresponding author
Additional information
Paarth Chothani and Vivek Mehta contributed equally to this work
Rights and permissions
About this article
Cite this article
Chothani, P., Mehta, V. & Stepanyants, A. Automated Tracing of Neurites from Light Microscopy Stacks of Images. Neuroinform 9, 263–278 (2011). https://doi.org/10.1007/s12021-011-9121-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-011-9121-2