Abstract
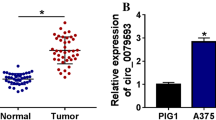
Circular RNAs (cicRNAs) have been identified to play pivotal roles in several cancer types. However, functions of circRNA in malignant melanoma are poor defined. Our current study demonstrated that human circMYC was obviously upregulated in human melanoma tissue. Furthermore, circMYC promoted the proliferation of human melanoma cells and Mel-CV cells. The expression of circMYC can repress Mel-CV cell glycolysis and LDHA activities in the in vitro glycolysis and lactate production evaluations. circMYC directly bound to miR-1236 as a molecular sponge that targeting miR-1236 in Mel-CV cells via bioinformatics analysis, pull-down assay, and luciferase reporter assays. Our present study revealed that 3′ UTR of LDHA acted as a target of miR-1236 using Mel-CV cells. Based on our findings, c-MYC-SRSF1 axis may regulate the production of circMYC. Overall, these results elucidate potential effects of circMYC in melanoma development and provide a promising biomarker for melanoma diagnosis.





Similar content being viewed by others
References
Siegel, R. L., Miller, K. D., & Jemal, A. (2018). Cancer statistics. CA: A Cancer Journal for Clinicians, 68, 7–30. https://doi.org/10.3322/caac.21442.
Mahadevan, A., Patel, V. L., & Dagoglu, N. (2015). Radiation therapy in the management of malignant melanoma. Oncology, 29, 743–751.
Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P., & Wu, M. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Molecular Cancer, 16, 94. https://doi.org/10.1186/s12943-017-0663-2.
Armakola, M., Higgins, M. J., Figley, M. D., Barmada, S. J., Scarborough, E. A., Diaz, Z., Fang, X., Shorter, J., Krogan, N. J., & Finkbeiner, S. (2012). Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nature Genetics, 44, 1302.
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., Zhong, G., Yu, B., Hu, W., & Dai, L. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nature Structural Molecular Biology, 22, 256.
Li, F., Zhang, L., Li, W., Deng, J., Zheng, J., An, M., Lu, J., & Zhou, Y. (2015). Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget, 6, 6001.
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., Evantal, N., Memczak, S., Rajewsky, N., & Kadener, S. (2014). circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell, 56, 55–66.
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., & Yang, B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research, 44, 2846–2858.
Pant, S., Hilton, H., & Burczynski, M. E. (2012). The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochemical Pharmacology, 83, 1484–1494.
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L., & Brown, P. O. (2013). Cell-type specific features of circular RNA expression. PLoS Genetics, 9, e1003777.
Wang, X., Zhang, Y., Huang, L., Zhang, J., Pan, F., Li, B., Yan, Y., Jia, B., Liu, H., & Li, S. (2015). Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. International Journal of Clinical and Experimental Pathology, 8, 16020.
Bian, D., Wu, Y., & Song, G. (2018). Novel circular RNA, hsa_circ_0025039 promotes cell growth, invasion and glucose metabolism in malignant melanoma via the miR-198/CDK4 axis. Biomedicine & Pharmacotherapy, 108, 165–176.
Luan, W., Shi, Y., Zhou, Z., Xia, Y., & Wang, J. (2018). circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochemical and Biophysical Research Communications, 502, 22–29.
Lin, X., Sun, R., Zhao, X., Zhu, D., Zhao, X., Gu, Q., Dong, X., Zhang, D., Zhang, Y., & Li, Y. (2017). C-myc overexpression drives melanoma metastasis by promoting vasculogenic mimicry via c-myc/snail/Bax signaling. Journal of Molecular Medicine, 95, 53–67.
Meyer, N., & Penn, L. Z. (2008). Reflecting on 25 years with MYC. Nature Reviews Cancer, 8, 976.
Kraehn, G., Utikal, J., Udart, M., Greulich, K., Bezold, G., Kaskel, P., Leiter, U., & Peter, R. (2001). Extra c-myc oncogene copies in high risk cutaneous malignant melanoma and melanoma metastases. British Journal of Cancer, 84, 72.
Nilsson, J. A., & Cleveland, J. L. (2003). Myc pathways provoking cell suicide and cancer. Oncogene, 22, 9007.
Wang, Y., Mo, Y., Gong, Z., Yang, X., Yang, M., Zhang, S., Xiong, F., Xiang, B., Zhou, M., & Liao, Q. (2017). Circular RNAs in human cancer. Molecular Cancer, 16, 25.
Gou, Q., Wu, K., Zhou, J.-K., Xie, Y., Liu, L., & Peng, Y. (2017). Profiling and bioinformatic analysis of circular RNA expression regulated by c-Myc. Oncotarget, 8, 71587.
Yang, Q., Du, W. W., Wu, N., Yang, W., Awan, F. M., Fang, L., Ma, J., Li, X., Zeng, Y., & Yang, Z. (2017). A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death and Differentiation, 24, 1609.
Yu, T., Wang, Y., Fan, Y., Fang, N., Wang, T., Xu, T., & Shu, Y. (2019). CircRNAs in cancer metabolism: a review. Journal of Hematology & Oncology, 12, 90.
Ruan, H., Xiang, Y., Ko, J., Li, S., Jing, Y., Zhu, X., Ye, Y., Zhang, Z., Mills, T., & Feng, J. (2019). Comprehensive characterization of circular RNAs in ~1000 human cancer cell lines. Genome Medicine, 11, 1–14.
Sunters, A., Armstrong, V. J., Zaman, G., Kypta, R. M., Kawano, Y., Lanyon, L. E., & Price, J. S. (2010). Mechano-transduction in osteoblastic cells involves strain-regulated estrogen receptor α-mediated control of insulin-like growth factor (IGF) I receptor sensitivity to ambient IGF, leading to phosphatidylinositol 3-kinase/AKT-dependent Wnt/LRP5 receptor-independent activation of β-catenin signaling. Journal of Biological Chemistry, 285, 8743–8758.
Panda A. C., Gorospe M. (2018). Detection and analysis of circular RNAs by RT-PCR. Bio Protocol, 8, e2775.
Gernapudi, R., Wolfson, B., Zhang, Y., Yao, Y., Yang, P., Asahara, H., & Zhou, Q. (2016). MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Molecular and Cellular Biology, 36, 30–38.
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., Sun, W., Dou, K., & Li, H. (2015). Circular RNA: a new star of noncoding RNAs. Cancer Letters, 365, 141–148.
Zdralevic, M., Brand, A., Di Ianni, L., Dettmer, K., Reinders, J., Singer, K., Peter, K., Schnell, A., Bruss, C., Decking, S. M., Koehl, G., Felipe-Abrio, B., Durivault, J., Bayer, P., Evangelista, M., O'Brien, T., Oefner, P. J., Renner, K., Pouyssegur, J., & Kreutz, M. (2018). Double genetic disruption of lactate dehydrogenases A and B is required to ablate the “Warburg effect” restricting tumor growth to oxidative metabolism. Journal of Biological Chemistry, 293, 15947–15961. https://doi.org/10.1074/jbc.RA118.004180.
Augoff, K., Hryniewicz-Jankowska, A., & Tabola, R. (2015). Lactate dehydrogenase 5: an old friend and a new hope in the war on cancer. Cancer Letters, 358, 1–7.
Sheng, S. L., Liu, J. J., Dai, Y. H., Sun, X. G., Xiong, X. P., & Huang, G. (2012). Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. The FEBS Journal, 279, 3898–3910.
Wang, Z.-Y., Loo, T. Y., Shen, J.-G., Wang, N., Wang, D.-M., Yang, D.-P., Mo, S.-L., Guan, X.-Y., & Chen, J.-P. (2012). LDH-A silencing suppresses breast cancer tumorigenicity through induction of oxidative stress mediated mitochondrial pathway apoptosis. Breast Cancer Research and Treatment, 131, 791–800.
Xie, H., Hanai J-i, Ren, J.-G., Kats, L., Burgess, K., Bhargava, P., Signoretti, S., Billiard, J., Duffy, K. J., & Grant, A. (2014). Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metabolism, 19, 795–809.
Ho, J., de Moura, M. B., Lin, Y., Vincent, G., Thorne, S., Duncan, L. M., Hui-Min, L., Kirkwood, J. M., Becker, D., & Van Houten, B. (2012). Importance of glycolysis and oxidative phosphorylation in advanced melanoma. Molecular Cancer, 11, 76.
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., & Kjems, J. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384.
An, J.-X., Ma, M.-H., Zhang, C.-D., Shao, S., Zhou, N.-M., & Dai, D.-Q. (2018). miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell International, 18, 66. https://doi.org/10.1186/s12935-018-0560-9.
Wang, Y., Yan, S., Liu, X., Zhang, W., Li, Y., Dong, R., Zhang, Q., Yang, Q., Yuan, C., & Shen, K. (2014). miR-1236-3p represses the cell migration and invasion abilities by targeting ZEB1 in high-grade serous ovarian carcinoma. Oncology Reports, 31, 1905–1910.
Dang, C. V. (2013). MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in Medicine, 3, a014217. https://doi.org/10.1101/cshperspect.a014217.
Das, S., Anczuków, O., Akerman, M., & Krainer, A. R. (2012). Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Reports, 1, 110–117.
Fischer G. M., Vashisht Gopal Y. N., McQuade J. L., Peng W., DeBerardinis R. J., Davies M. A. (2018). Metabolic strategies of melanoma cells: mechanisms, interactions with the tumor microenvironment, and therapeutic implications. Pigment Cell & Melanoma Research, 31, 11–30. https://doi.org/10.1111/pcmr.12661.
Parmenter, T. J., Kleinschmidt, M., Kinross, K. M., Bond, S. T., Li, J., Kaadige, M. R., Rao, A., Sheppard, K. E., Hugo, W., Pupo, G. M., Pearson, R. B., McGee, S. L., Long, G. V., Scolyer, R. A., Rizos, H., Lo, R. S., Cullinane, C., Ayer, D. E., Ribas, A., Johnstone, R. W., Hicks, R. J., & McArthur, G. A. (2014). Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discovery, 4, 423–433. https://doi.org/10.1158/2159-8290.CD-13-0440.
Funding
The study was supported by the Wuxi municipal health and Family Planning Commission Fund (MS201711).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Jin, C., Dong, D., Yang, Z. et al. CircMYC Regulates Glycolysis and Cell Proliferation in Melanoma. Cell Biochem Biophys 78, 77–88 (2020). https://doi.org/10.1007/s12013-019-00895-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-019-00895-0