Abstract
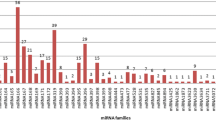
Withania somnifera, popularly known as Indian ginseng, is one of the most important medicinal plants. The plant is well studied in terms of its pharmaceutical activities and genes involved in biosynthetic pathways. However, not much is known about the regulatory mechanism of genes responsible for the production of secondary metabolites. The idea was to identify miRNA transcriptome responsible for the regulation of withanolide biosynthesis, specifically of root and leaf tissues individually. The transcriptome data of in vitro culture of root and leaf tissues of the plant was considered for miRNA identification. A total of 24 and 39 miRNA families were identified in root and leaf tissues, respectively. Out of these, 15 and 27 miRNA families have shown their involvement in different biological functions in root and leaf tissues, respectively. We report here, specific miRNAs and their corresponding target genes for corresponding root and leaf tissues. The target genes have also been analyzed for their role in withanolide metabolism. Endogenous root-miR5140, root-miR159, leaf-miR477, and leaf-miR530 were reported for regulation of withanolide biosynthesis.





Similar content being viewed by others
References
Kaileh, M., VandenBerghe, W., Boone, E., Essawi, T., & Haegeman, G. (2007). Screening of indigenous Palestinian medicinal plants for potential anti-inflammatory and cytotoxic activity. Journal of Ethnopharmacology, 113, 510–516.
Mirjalili, M. H., Moyano, E., Bonfill, M., Cusido, R. M., & Palazon, J. (2009). Steroidal lactones from Withania somnifera, an ancient plant for novel medicine. Molecules, 14(7), 2373–2393. https://doi.org/10.3390/molecules14072373
Siriwardane, A. S., Dharmadasa, R. M., & Samarasinghe, K. (2013). Distribution of withaferin A, an anticancer potential agent, in different parts of two varieties of Withania somnifera (L.) Dunal. grown in Sri Lanka. Pak J BiolSci, 16, 141–144.
Verma, S. K., & Kumar, A. (2011). Therapeutic uses of Withania somnifera (ashwagandha) with a note on withanolides and its pharmacological actions. Asian Journal of Pharmaceutical and Clinical Research, 4, 1–4.
Sangwan, R. S., Chaurasiya, N. D., Sangwan, P. L., Misra, L. N., Tuli, R., & Sangwan, N. S. (2008). Withanolide A is inherently de novo biosynthesized in roots of the medicinal plant Ashwagandha (Withania somnifera). Plant Physiology, 133(2), 278–287. https://doi.org/10.1111/j.1399-3054.2008.01076.x
Pal, S., Singh, S., Ashutosh, K. S., Madan, M. G., Suman, P. S. K., & Ajit, K. S. (2011). Comparative withanolide profiles, gene isolation, and differential gene expression in the leaves and roots of Withania somnifera. The Journal of Horticultural Science and Biotechnology, 86(4), 391–397. https://doi.org/10.1080/14620316.2011.11512779
Da Silva, A. C., Grativol, C., Thiebaut, F., Hemerly, A. S., & Ferreira, P. C. G. (2016). Computational identification and comparative analysis of miRNA precursors in three palm species. Planta, 243(5), 1265–1277. https://doi.org/10.1007/s00425-016-2486-6
Yang, T., Xue, L., & An, L. (2007). Functional diversity of miRNA in plants. Plant Science, 172(3), 423–432. https://doi.org/10.1016/j.plantsci.2006.10.009
Srivastava, S., Singh, N., Srivastava, G., & Sharma, A. (2017). miRNA mediated gene regulatory network analysis of Cichoriumintybus (chicory). Agri Gene, 3, 37–45. https://doi.org/10.1016/j.aggene.2016.11.003
Mallory, A. C., & Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nature Genetics, 38(Suppl), S31–S36. https://doi.org/10.1038/ng1791
Wang, J.-W., Wang, L.-J., Mao, Y.-B., Cai, W.-J., Xue, H.-W., & Chen, X.-Y. (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell, 17(8), 2204–2216. https://doi.org/10.1105/tpc.105.033076
Jones-Rhoades, M. W., & Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell, 14(6), 787–799. https://doi.org/10.1016/j.molcel.2004.05.027
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, 303(5666), 2022–2025. https://doi.org/10.1126/science.1088060
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., & Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature, 425(6955), 257–263. https://doi.org/10.1038/nature01958
Boke, H., Ozhuner, E., Turktas, M., Parmaksiz, I., Ozcan, S., & Unver, T. (2015). Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnology Journal, 13(3), 409–420. https://doi.org/10.1111/pbi.12346
Senthil, K., Jayakodi, M., Thirugnanasambantham, P., Lee, S. C., Duraisamy, P., Purushotham, P. M., Rajasekaran, K., Charles, S. N., Roy, I. M., Nagappan, A. K., Kim, G. S., Lee, Y. S., Natesan, S., Min, T. S., & Yang, T. J. (2015). Transcriptome analysis reveals in vitro cultured Withania somnifera leaf and root tissues as a promising source for targeted withanolide biosynthesis. BMC Genomics, 16, 1–16.
Patel, R. K., & Jain, M. (2012). NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One, 7(2), e30619. https://doi.org/10.1371/journal.pone.0030619
Kozomara, A., & Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 42(D1), D68–D73. https://doi.org/10.1093/nar/gkt1181
Numnark, S., Mhuantong, W., Ingsriswang, S., & Wichadakul, D. (2012). C-mii: a tool for plant miRNA and target identification. BMC Genomics, 13(Suppl 7), S16. https://doi.org/10.1186/1471-2164-13-S7-S16
Conesa, A., Gotz, S., García-Gómez, J.M., Terol, J., Talón, M. & Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18), 3674–3676
Botton, A., Galla, G., Conesa, A., Bachem, C., Ramina, A. & Barcaccia, G. (2008). Large-scale Gene Ontology analysis of plant transcriptome-derived sequences retrieved by AFLP technology. BMC Genomics, 9(1), 347
Galla, G., Barcaccia, G., Ramina, A., Collani, S., Alagna, F., Baldoni, L., Cultrera, N.G., Martinelli, F., Sebastiani, L. & Tonutti, P., (2009). Computational annotation of genes differentially expressed along olive fruit development. BMC Plant Biology, 9(1), 128
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. https://doi.org/10.1101/gr.1239303
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., & Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research, 35(Web Server), W182–W185. https://doi.org/10.1093/nar/gkm321
Singh, R., Pandey, N., Naskar, J., & Shirke, P. A. (2015). Physiological performance and differential expression profiling of genes associated with drought tolerance in contrasting varieties of two Gossypium species. Protoplasma, 252(2), 423–438. https://doi.org/10.1007/s00709-014-0686-0
Singh, P., Guleri, R., Angurala, A., Kaur, K., Kaur, K., Kaul, S. C., Wadhwa, R., & Pati, P. K. (2017). Addressing challenges to enhance the bioactives of Withania somnifera through organ, tissue, and cell culture based approaches. BioMed Research International, 2017, 1–15. https://doi.org/10.1155/2017/3278494
Dhar N, Razdan S, Rana S, Bhat WW, Vishwakarma R, Lattoo SK (2015) A decade of molecular understanding of withanolide biosynthesis and in vitro studies in Withania somnifera (L.) Dunal: prospects and perspectives for pathway engineering. Frontiers in Plant Science 6. doi:https://doi.org/10.3389/fpls.2015.01031.
Pal, S., Yadav, A. K., Singh, A. K., Rastogi, S., Gupta, M. M., Verma, R. K., Nagegowda, D. A., Pal, A., & Shasany, A. K. (2017). Nitrogen treatment enhances sterols and withaferin A through transcriptional activation of jasmonate pathway, WRKY transcription factors, and biosynthesis genes in Withania somnifera (L.) Dunal. Protoplasma, 254(1), 389–399. https://doi.org/10.1007/s00709-016-0959-x
Davis, L., & Kuttan, G. (2000). Immunomodulatory activity of Withania somnifera. Journal of Ethnopharmacology, 71(1-2), 193–200. https://doi.org/10.1016/S0378-8741(99)00206-8
Khanna, D., Sethi, G., Ahn, K. S., et al. (2007). Natural products as a gold mine for arthritis treatment. CurrOpinPharmacol, 7, 344–351.
Ku, S. K., & Bae, J. S. (2014). Antiplatelet, anticoagulant, and profibrinolytic activities of withaferin A. VascPharmacol, 60, 120–126.
Datta, A., Bagchi, C., & Tripathi, S. K. (2013). Antidiabetic and antihyperlipidemic activity of hydroalcoholic extract of Withania coagulans Dunal dried fruit in experimental rat models. J Ayurveda Integr Med, 4, 99–106.
Choudhary, S., Kumar, P., & Malik, J. (2013). Plants and phytochemicals for Huntington’s disease. Pharmacognosy Reviews, 7(14), 81–91. https://doi.org/10.4103/0973-7847.120505
Pingali, U., Pilli, R., & Fatima, N. (2014). Effect of standardized aqueous extract of Withania somnifera on tests of cognitive and psychomotor performance in healthy human participants. Pharmacogn Res, 6(1), 12–18. https://doi.org/10.4103/0974-8490.122912
Rai, M., Jogee, P. S., Agarkar, G., & dos Santos, C. A. (2016). Anticancer activities of Withania somnifera: current research, formulations, and future perspectives. Pharmaceutical Biology, 54(2), 189–197. https://doi.org/10.3109/13880209.2015.1027778
Griffiths, J. S., Saini, H. K., Van, D. S., & Enright, A. J. (2008). miRBase: tools for microRNA genomics, Nucleic Acids Res., 36, D154eD158.
Xu, Y., Chu, L., Jin, Q., Wang, Y., Chen, X., Zhao, H., & Xue, Z. (2015). Transcriptome-wide identification of miRNAs and their targets from Typhaangustifolia by RNA-Seq and their response to cadmium stress. PLoS One, 10(4), e0125462. https://doi.org/10.1371/journal.pone.0125462
Saifi, M., Nasrullah, N., Ahmad, M. M., Ali, A., Khan, J. A., & Abdin, M. Z. (2015). In silico analysis and expression profiling of miRNAs targeting genes of steviol glycosides biosynthetic pathway and their relationship with steviol glycosides content in different tissues of Stevia rebaudiana. Plant Physiology and Biochemistry, 94, 57–64. https://doi.org/10.1016/j.plaphy.2015.05.009
Koul, A., Yogindran, S., Sharma, D., Kaul, S., Rajam, M. V., & Dhar, M. K. (2016). Carotenoid profiling, in silico analysis and transcript profiling of miRNAs targeting carotenoid biosynthetic pathway genes in different developmental tissues of tomato. Plant Physiology and Biochemistry, 108, 412–421. https://doi.org/10.1016/j.plaphy.2016.08.001
Sharma, D., Tiwari, M., Pandey, A., Bhatia, C., Sharma, A., & Trivedi, P. K. (2016). MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development in Arabidopsis. Plant Physiology, pp.01831.2015. https://doi.org/10.1104/pp.15.01831
Gupta, O. P., Karkute, S. G., Banerjee, S., Meena, N. L., & Dahuja, A. (2017). Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Frontiers in Plant Science, 8. https://doi.org/10.3389/fpls.2017.00374
Bologna, N. G., Mateos, J. L., Bresso, E. G., & Palatnik, J. F. (2009). A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. The EMBO Journal, 28(23), 3646–3656. https://doi.org/10.1038/emboj.2009.292
Bologna, N. G., Schapire, A. L., Zhai, J., Chorostecki, U., Boisbouvier, J., Meyers, B. C., & Palatnik, J. F. (2013). Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Research, 23(10), 1675–1689. https://doi.org/10.1101/gr.153387.112
Das, A., Das, P., Kalita, M. C., & Mondal, T. K. (2016). Computational identification, target prediction, and validation of conserved miRNAs in insulin plant (Costuspictus D. Don). Applied Biochemistry and Biotechnology, 178(3), 513–526. https://doi.org/10.1007/s12010-015-1891-9
Xu, S., Jiang, Y., Wang, N., Xia, B., Jiang, Y., Li, X., Zhang, Z., Li, Y., & Wang, R. (2016). Identification and differential regulation of microRNAs in response to methyl jasmonatetreatment in Lycorisaurea by deep sequencing. BMC Genomics, 17(1), 789. https://doi.org/10.1186/s12864-016-2645-y
Xu, W., Cui, Q., Li, F., & Liu, A. (2013). Transcriptome-wide identification and characterization of microRNAs from castor bean (Ricinuscommunis L.) PLoS One, 8(7), e69995. https://doi.org/10.1371/journal.pone.0069995
Zhang, B. H., Pan, X. P., Cox, S. B., Cobb, G. P., & Anderson, T. A. (2006). Evidence that miRNAs are different from other RNAs. Cellular and Molecular Life Sciences CMLS, 63(2), 246–254. https://doi.org/10.1007/s00018-005-5467-7
Yang, Y., Chen, X., Chen, J., Xu, H., Li, J., & Zhang, Z. (2011). Differential miRNA expression in Rehmanniaglutinosa plants subjected to continuous cropping. BMC Plant Biology, 11(1), 53. https://doi.org/10.1186/1471-2229-11-53
Kaur, P., Shukla, N., Joshi, G., VijayaKumar, C., Jagannath, A., Agarwal, M., Goel, S., & Kumar, A. (2017). Genome-wide identification and characterization of miRNAome from tomato (Solanum lycopersicum) roots and root-knot nematode (Meloidogyne incognita) during susceptible interaction. PLoS One, 12(4), e0175178. https://doi.org/10.1371/journal.pone.0175178
Lu, Y.-B., Qi, Y.-P., Yang, L.-T., Guo, P., Li, Y., & Chen, L.-S. (2015). Boron-deficiency-responsive microRNAs and their targets in Citrus sinensis leaves. BMC Plant Biology, 15(1). https://doi.org/10.1186/s12870-015-0642-y
Dutt, S., Manjul, A. S., Raigond, P., Singh, B., Siddappa, S., Bhardwaj, V., Kawar, P. G., Patil, V. U., & Kardile, H. B. (2017). Key players associated with tuberization in potato: potential candidates for genetic engineering. Critical Reviews in Biotechnology, 1–19(7), 942–957. https://doi.org/10.1080/07388551.2016.1274876
Miozzi, L., Napoli, C., Sardo, L., & Accotto, G. P. (2014). Transcriptomics of the interaction between the monopartite phloem-limited geminivirus tomato yellow leaf curl Sardinia virus and Solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during infection. PLoS One, 9(2), e89951. https://doi.org/10.1371/journal.pone.0089951
Khaldun, A. B. M., Huang, W., Liao, S., Lv, H., & Wang, Y. (2015). Identification of MicroRNAs and target genes in the fruit and shoot tip of Lyciumchinense: a traditional Chinese medicinal plant. PLoS One, 10(1), e0116334. https://doi.org/10.1371/journal.pone.0116334
Gao, C., Wang, P., Zhao, S., Zhao, C., Xia, H., Hou, L., Ju, Z., Zhang, Y., Li, C., & Wang, X. (2017). Small RNA profiling and degradome analysis reveal regulation of microRNA in peanut embryogenesis and early pod development. BMC Genomics, 18(1), 220. https://doi.org/10.1186/s12864-017-3587-8
Song, C., Yu, M., Han, J., Wang, C., Liu, H., Zhang, Y., & Fang, J. (2012). Validation and characterization of Citrus sinensis microRNAs and their target genes. BMC Research Notes, 5(1), 235. https://doi.org/10.1186/1756-0500-5-235
Omidvar, V., Mohorianu, I., Dalmay, T., & Fellner, M. (2015). Identification of miRNAs with potential roles in regulation of anther development and male-sterility in 7B-1 male-sterile tomato mutant. BMC Genomics, 16(1). https://doi.org/10.1186/s12864-015-2077-0
Liang, G., Li, Y., He, H., Wang, F., & Yu, D. (2013). Identification of miRNAs and miRNA-mediated regulatory pathways in Carica papaya. Planta, 238(4), 739–752. https://doi.org/10.1007/s00425-013-1929-6
Mangrauthia, S. K., Bhogireddy, S., Agarwal, S., Prasanth, V. V., Voleti, S. R., Neelamraju, S., & Subrahmanyam, D. (2017). Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. Journal of Experimental Botany, 68(9), 2399–2412. https://doi.org/10.1093/jxb/erx111
Campbell, J. A., Davies, G. J., Bulone, V., & Henrissat, B. (1997). A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. The Biochemical Journal, 326(3), 929–939. https://doi.org/10.1042/bj3260929
Jones, P., & Vogt, T. (2001). Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta, 213(2), 164–174. https://doi.org/10.1007/s004250000492
Bhat, W. W., Lattoo, S. K., Razdan, S., Dhar, N., Rana, S., Dhar, R. S., Khan, S., & Vishwakarma, R. A. (2012). Molecular cloning, bacterial expression and promoter analysis of squalene synthase from Withania somnifera (L.) Dunal. Gene, 499(1), 25–36. https://doi.org/10.1016/j.gene.2012.03.004
Chen, X., Dai, G., Ren, Z., Tong, Y., Yang, F., & Zhu, Y. (2016). Identification of dietetically absorbed rapeseed (Brassica campestris L.) bee pollen microRNAs in serum of mice. BioMed Research International, 2016, 1–5. https://doi.org/10.1155/2016/5413849
Chin, A. R., Fong, M. Y., Somlo, G., Wu, J., Swiderski, P., Wu, X., & Wang, S. E. (2016). Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Research, 26(2), 217–228. https://doi.org/10.1038/cr.2016.13
Liang, G., Zhu, Y., Sun, B., Shao, Y., Jing, A., Wang, J., & Xiao, Z. (2014). Assessing the survival of exogenous plant microRNA in mice. Food Science & Nutrition, 2(4), 380–388. https://doi.org/10.1002/fsn3.113
Acknowledgements
We would like to thank SRA database of NCBI database for making it freely available to the scientific community. Financial assistance to SS under BTISnet programme of DBT-New Delhi, S and RS under SERB-PDF and GS under ICMR-SRF is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Figure 1
(DOCX 892 kb)
Supplementary Figure 2
(DOCX 583 kb)
Supplementary Figure 3
(DOCX 507 kb)
Supplementary Figure 4
(DOCX 502 kb)
Supplementary Table 1
(DOCX 16 kb)
Supplementary Table 2
(DOCX 36 kb)
Supplementary Table 3
(DOCX 15 kb)
Supplementary Table 4
(DOCX 30 kb)
Supplementary Table 5
(DOCX 58 kb)
Rights and permissions
About this article
Cite this article
Srivastava, S., Sanchita, Singh, R. et al. Comparative Study of Withanolide Biosynthesis-Related miRNAs in Root and Leaf Tissues of Withania somnifera. Appl Biochem Biotechnol 185, 1145–1159 (2018). https://doi.org/10.1007/s12010-018-2702-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-018-2702-x