Abstract
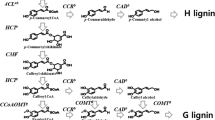
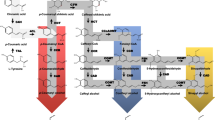
Sugarcane (Saccharum spp.) is one of the highest biomass-producing plant and the best lignocellulosic feedstock for ethanol production. To achieve more efficient conversion of biomass to ethanol, a better understanding of the main factors affecting biomass recalcitrance is needed. Therefore, with this objective, here, we report a systematic study on lignin content, deposition, identification, and cloning of genes involved in lignin biosynthesis and their differential expression in five sugarcane clones, EC11003, EC11010, IK 76-91, IK 76-99, and Co 86032. Lignin content among the clones varied from 26.87 to 23.19 % with the highest in the clone EC11010 and the lowest in high sugar Co86032. Lignin deposition studied through phloroglucinol staining of the cell walls implied that the sclerenchyma cells of the energy canes (EC11010 and EC11003) have more lignin deposition followed by the Erianthus (IK 76-91 and IK 76-99) clones whereas Co86032 has the minimum amount of lignin deposition. We cloned partial coding regions of important genes of lignification COMT (650 bp), CCR (332 bp), and PAL (650 bp) from Erianthus, wild relative of sugarcane followed by the expression analysis through real-time PCR. Differential expression analysis showed high level of expression for the three genes in the energy cane EC11010.






Similar content being viewed by others
References
Canilha, L., Chandel, A. K., Milessi, T. S dos S, Antunes, F. A. F., Freitas, C. da. L. W., Felipe, A. G. das. M., Silva, S. da. S. S. (2012). Bioconversion of sugarcane biomass into ethanol: an overview about composition, pretreatment methods, detoxification of hydrolysates, enzymatic saccharification, and ethanol fermentation. Journal of Biomedicine and Biotechnology, 15 pages.
Annual Report Sugarcane breeding Institute. 2011-12.
Chen, F., & Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nature Biotechnology, 25, 759–761.
Casu, R. E., Dimmock, C. M., Chapman, S. C., Grof, C. P., McIntyre, C. L., Bonnett, G. D., & Manners, J. M. (2004). Identification of differentially expressed transcripts from maturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Molecular Biology, 54, 503–517.
Van Acker, R., Vanholme, R., Storme, V., Mortimer, J. C., Dupree, P., & Boerjan, W. (2013). Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnology for Biofuels, 6, 46.
Ramos, R. L., Tovar, F. J., Junqueira, R. M., Lino, F. B., & Sachetto-Martins, G. (2001). Sugarcane expressed sequences tags (ESTs) encoding enzymes involved in lignin biosynthesis pathways. Genetics and Molecular Biology, 24, 235–241.
Papini-Terzi, F. S., Rocha, F. R., Vêncio, R. Z., Oliveira, K. C., Felix, J. d. M., Vicentini, R., Rocha, C. d. S., Simões, A. C., Ulian, E. C., & di Mauro, S. M. (2005). Transcription profiling of signal transduction-related genes in sugarcane tissues. DNA Research, 12, 27–38.
Lakshmi, K., & Kalaivani, A. (2015). Molecular cloning of cDNA from sugarcane coding for caffeic acid O-methyltransferase (COMT) involved in lignification. SugarTech. doi:10.1007/s12355-015-0417-7.
Bottcher, A., Cesarino, I., Santos, A. B., Vicentini, R., Mayer, J. L., Vanholme, R., et al. (2013). Lignification in sugarcane: biochemical characterization, gene discovery, and expression analysis in two genotypes contrasting for lignin content. Plant Physiology, 163(4), 539–1557.
Hahlbrock, K., & Scheel, D. (1989). Physiology and molecular biology of phenylpropanoid metabolism. Annual Reviews of Plant Physiology and Plant Molecular Biology, 40, 347–369.
Dixon, R. A., & Lamb, C. J. (1990). Molecular communication in interactions between plants and microbial pathogens. Annual Review of Plant Physiology and Plant Molecular Biology, 41, 339–367.
Sluiter, A., Hames, B., Ruiz, R., Scarlata, C., Sluiter, J., Templeton, D., Crocker D. (2011) Determination of structural carbohydrates and lignin in biomass laboratory analytical procedure (LAP) Issue Date: April 2008 Revision Date: July, Version 07–08- 2011.
Steve, R, Skaletsky, H. J. (1998). Primer3. Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html.
Johansen, D. A. (1940). Plant microtechnique McGraw Hill book Co. New York 523.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods, 25, 402–408.
Hatfield, R. D., Marita, J. M., Frost, K., Grabber, J., Ralph, J., Fachuang, L., & Hoon, K. (2009). Grass lignin acylation:p-coumaroyl transferase activity and cell wall characteristics of C3 and C4 grasses. Planta, 229, 1253–1267.
Masarin, F., Gurpilhares, D. B., Baffa, D. C. F., Barbosa, M. H. P., Carvalho, W., Ferraz, A., & Milagres, A. M. F. (2011). Chemical composition and enzymatic digestibility of sugarcane selected for varied lignin content. Biotechnology of Biofuels, 4, 55.
SanJuan, R., Anzaldo, J., Vargas, J., Turrado, J., & Patt, R. (2001). Morphological and chemical composition of pitch and fibers from Mexican sugarcane bagasse. Holz als Roh-und Werkstoff, 59, 447–450.
Joshi, C. P., & Chiang, V. L. (1998). Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Molecular Biology, 37, 663–674.
Guillaumie, S., Goffner, D., Barbier, O., Martinant, J. P., Pichon, M., & Barrière, Y. (2008). Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT); down-regulated, and normal maize plants. BMC Plant Biology, 8, 71.
Ferrer, J. L., Austin, M. B., Stewart, C., & Noel, J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiology and Biochemistry, 46(3), 356–370.
Jornvall, H., Persson, B., Krook, M., Atrian, S., Gonzalez-Duarte, R., Jeffery, J., & Ghosh, D. (1995). Short-chain dehydrogenases/reductases (SDR). Biochemistry, 34, 6003–6013.
Barakat, A., Yassin, N. B. M., Park, J. S., Choi, A., Herr, J., & Carlson, J. E. (2011). Comparative and phylogenomic analyses of cinna-moyl-CoA reductase and cinnamoyl-CoA-reductase-like gene family in land plants. Plant Science, 181(3), 249–257.
Hua, C., Linling, L., & Shuiyuan, C. (2012). Characterization of a cinnamoyl-CoA reductase gene in Ginkgo biloba: effects on lignification and environmental stresses. African Journal of Biotechnology, 11(26), 6780–6794.
Chen, L., Auh, C., Chen, F., Cheng, X., Aljoe, H., Dixon, R. A., & Wang, Z. (2002). Lignin deposition and associated changes in anatomy, enzyme activity, gene expression, and ruminal degradability in stems of tall fescue at different developmental stages. Journal of Agriculture and Food Chemistry, 50, 5558–5565.
Cesarino, I., Araujo, P., Sampaio Mayer, J. L., Paes Leme, A. F., & Mazzafera, P. (2012). Enzymatic activity and proteomic profile of class III peroxidases during sugarcane stem development. Plant Physiology and Biochemistry, 55, 66–76.
Tamasloukht, B., Wong Quai Lam, M. S., Martinez, Y., Tozo, K., Barbier, O., Jourda, C., Jauneau, A., Borderies, G., Balzergue, S., Renou, J. P., Huguet, S., Martinant, J. P., Tatout, C., Lapierre, C., Barriere, Y., Goffner, D., & Pichon, M. (2011). Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: effects on lignification, fiber development, and global gene expression. Journal of Experimental Botany, 62, 3837–3848.
Zhao, J., Davis, C. L., & Verpoortec, R. (2005). Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnology Advances, 23(4), 283–333.
Hashemitabar, M., Kolahi, M., Tabandeh, R. M., Jonoubi, P., & Majd, A. (2014). cDNA cloning, phylogenic analysis and Gene expression pattern of phenylalanine ammonia-lyase in sugarcane (Saccharum officinarum L.). Brazilian Archives of Biology and Technology, 57(4), 456–465.
Zhang, Q., Cheetamun, R., Dhugga, K. S., Rafalski, J. A., Tingey, S. V., Shirley, N. J., Taylor, J., Hayes, K., Beatty, M., Bacic, A., Burton, R. A., & Fincher, G. B. (2014). Spatial gradients in cell wall composition and transcriptional profiles along elongating maize internodes. BMC Plant Biology, 14, 27.
Hu, Y., Di, P., Chen, J., Xiao, Y., Zhang, L., & Chen, W. (2011). Isolation and characterization of a gene encoding cinnamoyl-CoA reductase from Isatis indigotica fort. Molecular Biology Reports, 38(3), 2075–2083.
Koutaniemi, S. T., Warinowski, A. K., & Arkonen (2007). Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real-time RT-PCR. Plant Molecular Biology, 65(3), 311–328.
Acknowledgments
This work has been supported by the DST SERB project No. SB/EMEQ-001/2013. The authors wish to thank Dr. Bakshi Ram, Director and Dr. M. N. Premachandran, Head, Division of Crop Improvement, Sugarcane Breeding Institute, Coimbatore, for kindly providing genetically pure clones of the sugarcane hybrids for carrying out the work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kasirajan, L., Aruchamy, K., Thirugnanasambandam, P.P. et al. Molecular Cloning, Characterization, and Expression Analysis of Lignin Genes from Sugarcane Genotypes Varying in Lignin Content. Appl Biochem Biotechnol 181, 1270–1282 (2017). https://doi.org/10.1007/s12010-016-2283-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2283-5