Abstract
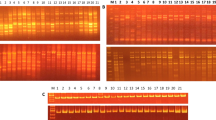
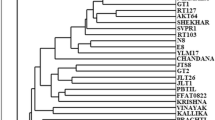
In order to compare the effectiveness of two molecular marker systems, a set of six RAPD and nine SSR markers were used to study the genetic diversity in a population of 46 sour orange accessions, a common rootstock used in almost all citrus orchards in Tunisia. Genetic diversity parameters [average and effective number of alleles, percentage of polymorphism, polymorphic information content (PIC), effective marker index (EMI), and marker index (MI) parameters] for RAPD, SSR, and RAPD + SSR were determined in order to assess the efficiency of the two marker systems. The results revealed that these parameters were significantly higher when using RAPD markers. Similarly, cluster analysis using the results of RAPD was practically the same as that obtained when combining data from the two marker systems (RAPD + SSR) demonstrating the efficiency of RAPD in discriminating between sour orange accessions. Therefore, the use of SSR markers, known to be more efficient and discriminatory, does not bring significant supplementary information in this work. Indeed, results would have been obtained using only the RAPD markers. Accordingly, this work highlights the efficiency and advantages of RAPD, as an easy and efficient technique, in studying citrus rootstock’s genetic diversity, and establishing genetic relationships among citrus accessions.




Similar content being viewed by others
References
Abkenar, A. A., & Isshiki, S. (2003). Molecular characterization and genetic diversity among Japanese acid citrus (Citrus spp.) based on RAPD markers. J Hort Sci Biotech, 78(1), 108–112.
Aka-Kacar, Y., Demirel, A., Tuzcu, O., Yesiloglu, T., et al. (2005). Preliminary results on fingerprinting lemon genotypes tolerant to mal secco (Phoma tracheiphila Kanc. et Ghik) disease by RAPD markers. Biologia, 60, 295–300.
Bachmann, K. (1997). Nuclear DNA markers in plant biosystematicresearch. Opera Botanica, 132, 137–148.
Baraket, G., Chatti, K., Saddoud, O., Abdelkarim, A. B., Mars, M., Trifi, M., & Hannachi, A. S. (2010). Comparative assessment of SSR and AFLP markers for evaluation of genetic diversity and conservation of fig, Ficus carica L., genetic resources in Tunisia. Plant Molecular Biology Reporter, 29, 171–184.
Bar-Joseph, M., Marcus, R., & Lee, R. F. (1989). The continuous challenge of citrus tristeza virus control. Annual Review of Phytopathology, 27, 291–316.
Barkley, N., Roose, M., Krueger, R., & Frederici, C. (2006). Assessing genetic diversity and population structure in a Citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theoretical and Applied Genetics, 112, 1519–1531.
Barkley, N. A., Krueger, R. R., Federici, C. T., & Roose, M. L. (2009). What phylogeny and gene genealogy analyses reveal about homoplasy in citrus microsatellite alleles. Plant Systematics and Evolution, 282, 71–86.
Barrett, H. C., & Rhodes, A. M. (1976). A numerical taxonomic study of affinity relations in cultivated citrus and its close relatives. Systematic Botany, 1, 105–136.
Belaj, A., Satovic, Z., Cipriani, G., Baldoni, L., Testolin, R., Rallo, L., & Trujillo, I. (2003). Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theoretical and Applied Genetics, 107, 736–744.
Bernardo Royo, J., & Itoiz, R. (2004). Evaluation of the discriminance capacity of RAPD, isoenzymes and morphologic markers in apple (Malus × domestica Borkh.) and the congruence among classifications. Genetic Resources and Crops Evolution, 51, 153–160.
Biswas, M. K., Xu, Q., & Deng, X. X. (2010). Utility of RAPD, ISSR, IRAP and REMAP markers for the genetic analysis of Citrus spp. Scientia Horticulturae, 124, 254–261.
Cai, Q., Guy, C. L., & Moore, G. A. (1994). Extension of the linkage map in Citrus using random amplified polymorphic DNA (RAPD) marker and RFLP mapping of cold-acclimation-responsive loci. Theoretical and Applied Genetics, 89, 606–614.
Callen, D. F., Callen, A. D., Thimpson, Y., Shen, H. A., Phillips, R. I., Richards, J. C., & Mulley, G. R. (1993). Sutherland incidence and origin of “null” alleles in the (AC)n microsatellite markers. American Journal of Human Genetics, 52, 922–927.
Campos, T. E., Espinosa, M. A. G., Warburton, M. L., Varela, A. S., et al. (2005). Characterization of mandarin (Citrus spp.) using morphological and AFLP markers. Interciencia, 30, 687–693.
Capparelli, R., Viscardi, M., Amoroso, M. G., Blaiotta, G., & Bianco, M. (2004). Inter-simple sequence repeat markers and flow cytometry for the characterization of closely related Citrus limon germplasm. Biotechnology Letters, 26, 1296–1299.
Castle, W. S., Tucker, D. P. H., Krezdorn, A. H., & Youtsey, C. O. (1993). Rootstocks for Florida citrus: rootstock selection, the first step to success (2nd ed.). Gainesville: University of Florida. 92p.
Cipriani, G., Di Bella, R., & Testolin, R. (1996). Screening RAPD primers for molecular taxonomy and cultivar fingerprinting in the genus Actinidia. Euphytica, 90, 169–174.
Devarumath, R. M., Doule, R. B., Kawar, P. G., Naikebawane, S. B., & Nerkar, Y. S. (2007). Field performance and RAPD analysis to evaluate genetic fidelity of tissue culture raised plants vis à vis conventional sets derived plants of sugarcane. Sugar Tech, 9(1), 17–22.
Dice, L. R. (1945). Measures of the amount of ecologic association between species. Ecology, 26, 297–302.
Elisiario, P. J., Justo, E. M., & Leitao, J. M. (1999). Identification of mandarin hybrids by isozyme and RAPD analysis. Scientia Horticulturae, 81, 287–299.
Esselman, E. J., Lianqiang, L., Crawford, D. J., Winduss, J. L., et al. (1999). Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Molecular Ecology, 8, 443–451.
Fang, D. Q., & Roose, M. L. (1997). Identification of closely related Citrus cultivars with intersimple sequence repeat markers. Theoretical and Applied Genetics, 95, 408–417.
Fang, D. Q., Krueger, R. R., & Roose, M. L. (1998). Phylogenetic relationships among selected Citrus germplasm accessions revealed by inter-simple sequence repeat (ISSR) markers. Journal of the American Society for Horticultural, 123, 612–617.
FAOSTAT (2012). FAO Statistics Database on the World Wide Web. http://faostat.fao.org/site/567/default.aspx#ancor (accessed August 2012).
Fernandez, M. E., Figueras, A. M., & Benito, C. (2002). The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification and genetic diversity among barley cultivars with known origin. Theoretical and Applied Genetics, 104, 845–851.
Fu, C. H., Chen, C. L., Guo, W. W., & Deng, X. X. (2004). GISH, AFLP and PCR-RFLP analysis of an intergeneric somatic hybrid combining Goutou sour orange and Poncirus trifoliata. Plant Cell Reports, 23, 391–396.
Gallego, F. J., Perez, M. A., Nunez, Y., & Hidalgo, P. (2005). Comparison of RAPDs, AFLPs and SSR markers for the genetic analysis of yeast strains of Saccharomyces cerevisiae. Food Microbiology, 22, 561–568.
Garcia, A. A. F., Benchimol, L. L., Barbosa, A. M. M., Geraldi, I. O., et al. (2004). Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genetics and Molecular Biology, 27, 579–588.
Gilbert, J. E., Lewis, R. V., Wilkinson, M. J., & Caligari, P. D. (1999). Developing an appropriate strategy to assess genetic variability in plant collections. Theoretical and Applied Genetics, 98, 1125–1131.
Graner, A., Dehmer, K. J., Thiel, T., & Börner, A. (2004). Plant genetic resources: benefits and implications of using molecular markers. In M. C. de Vincente (Ed.), Issues in genetic resources no. 11 (pp. 26–32). Rome: IPGRI.
Guilford, P., Prakash, S., Zhu, J. M., Rikkerink, E., Gardiner, S., Bassett, H., & Forster, R. (1997). Microsatellites in Malus domestica (apple): abundance, polymorphism and cultivar identification. Theoretical and Applied Genetics, 94, 249–254.
Gulsen, O., & Roose, M. L. (2001). Lemons: diversity and relationships with selected citrus genotypes as measured with nuclear genome markers. Journal of the American Society for Horticultural Science, 126, 309–317.
Gulsen, O., Uzun, A., Canan, I., Seday, U., & Canihos, E. (2010). A new citrus linkage map based on SRAP, SSR, ISSR, POGP, RGA and RAPD markers. Euphytica, 173, 265–277.
Gupta, P. K., Varshney, R. K., & Prasad, M. (2002). Molecular markers: principles and methodology. In S. M. Jain, B. S. Ahloowalia, & D. S. Brar (Eds.), Molecular techniques in crop improvement (pp. 9–54). The Netherlands: Kluwer Academic Publishers.
Jaccard, P. (1908). Nouvelles recherché sur la distribution florale. Bulletin de la Société vaudoise des sciences naturelles , 44, 223–270.
Kalinowski, S. T., Taper, M. L., & Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 16, 1099–1106. doi:10.1111/j.1365-294x.2007.03089.x.
Khadari, B., Breton, C., Moutier, N., Roger, J. P., Besnard, G., Bervillé, A., et al. (2003). The use of molecular markers for germplasm management in a French olive collection. Theoretical and Applied Genetics, 106, 521–529.
Landergott, U., Holderegger, R., Kozlowski, G., & Schneller, J. J. (2001). Historical bottlenecks decrease genetic diversity in natural populations of Dryopteris cristata. Heredity, 87, 344–355.
Luro, F., Costantino, G., Terol, J., Argout, X., Allario, T., Wincker, P., Talon, M., Ollitrault, P., & Morillon, R. (2008). Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics, 9, 287.
Luro, F., Rist, D., & Ollitrault, P. (2001). Evaluation of genetic relationships in Citrus genus by means of sequence tagged microsatellites. Proceedings of the International Symposium on Molecular Markers for Characterizing Genotypes and Identifying Cultivars in Horticulture February 2001; Montpellier, France. Acta Horticulturae, 546, 237–242.
Lynch, M., & Walsh, J. B. (1998). Genetics and analysis of quantitative traits. Sunderland: Sinauer Assocs., Inc.
Mabberley, D. J. (1998). Australian Citreae with notes on other Aurantioideae (Rutaceae). Telopea, 74, 333–344.
Roose, M. L. (http://www.plantbiology.ucr.edu/faculty/SSR-Marker-Tables-Germplasm-v5.pdf).
Ming, F., Liu, Q. K., Shi, J. L., Wang, W., & Lu, B. R. (2009). Strategic conservation of orchard germplasm based on indigenous knowledge and genetic diversity: a case study of sour orange populations in China. Journal of Integrative Plant Biology, 51, 100–106.
Morgante, M., & Olivieri, A. M. (1993). PCR-amplified microsatellite as markers in plant genetics. Plant Journal, 3, 175–182.
Nicese, F. P., Hormaza, J. I., & McGranahan, G. H. (1998). Molecular characterization and genetic relatedness among walnut (Juglans regia L.) genotypes based on RAPD markers. Euphytica, 101, 199–206.
Penner, G. A. (1996). RAPD analysis of plant genomes. In P. P. Jauhar (Ed.), Methods of genome analysis in plants (pp. 251–268). Boca Raton: CRC Press.
Nicolosi, E., Deng, Z. N., Gentile, A., & La Malfa, S. (2000). Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theoretical and Applied Genetics, 100, 1155–1166.
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Research, 27, 209–220.
Novelli, V. M., Cristofani, M., Souza, A. A., & Machado, M. A. (2006). Development and characterization of polymorphic microsatellite markers for the sweet orange (Citrus sinensis L. Osbeck). Genetics and Molecular Biology, 29, 90–96.
Novelli, V. M., Machado, M. A., & Romero Lopes, C. (2000). Isoenzymatic polymorphism in Citrus spp. and Poncirus trifoliata (L.) Raf. (Rutaceae). Genetics and Molecular Biology, 23, 163–168.
Oliveira, R. P., Cristofani, M., Aguilar-vildoso, C. I., & Machado, M. A. (2002). Diversidade genética entre híbridos de tangerina ‘Cravo’ e laranja ‘Pêra’. Pesquisa Agropecuária Brasileira, Brasília, 37(4), 479–484.
Ollitrault, F., Terol, J., Pina, J. A., Navarro, L., Talon, M., & Ollitrault, P. (2010). Development of SSR markers from Citrus clementina (Rutaceae) BAC end sequences and interspecific transferability in Citrus. American Journal of Botany, 97, e124–e129.
Palombi, M. A., & Damiano, C. (2002). Comparison between RAPD and SSR molecular mark- ers in detecting genetic variation in kiwifruit (Actinidia deliciosa A. Chev). Plant Cell Reports, 20, 1061–1066.
Pejic, I., Ajmone-Marsan, P., Morgante, M., Kozumplick, V., Castiglioni, P., Taramino, G., & Motto, M. (1998). Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs, and AFLPs. Theoretical and Applied Genetics, 97, 1248–1255.
Pooler, M., & Scorza, R. (1995). Aberrant transmission of RAPD markers in haploids, doubled haploids, and F1 hybrids of peach: observations and speculation on causes. Scientia Horticulturae, 64, 233–241.
Powell, W., Morgante, M., Andre, C., Hanafey, M., Vogel, J., Tingey, S., & Rafalski, A. (1996). The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding, 2, 225–238.
Prevost, A., & Wilkinson, M. J. (1999). A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theoretical and Applied Genetics, 98, 107–112.
Rajora, O. P., & Rahman, M. H. (2003). Microsatellite DNA and RAPD fingerprinting, identification and genetic relationships of hybrid poplar (Populus canadiensis) cultivars. Theoretical and Applied Genetics, 106, 470–477.
Rohlf, F. J. (2000). NTSYS- pc: Numerical taxonomy and multivariate analysis system. Version: 2.1. Exeter Software, New York.
Ruiz, C., Paz Breto, M., & Asíns, M. J. (2000). A quick methodology to identify sexual seedlings in citrus breeding programs using SSR markers. Euphytica, 112, 89–94.
Rus-Kortekaas, W., Smulders, M. J. M., Arens, P., & Vosman, B. (1994). Direct comparison of levels of genetic-variation in tomato detected by a GACA containing microsatellite probe and by random amplified polymorphic DNA. Genome, 37, 375–381.
Russell, J. R., Fuller, J. D., & Macaulay, M. (1997). Direct comparison of levels of genetic variation among barley accessions detected by RFLPs, AFLPs, SSRs and RAPDs. Theoretical and Applied Genetics, 95, 714–722.
Scariot, V., De Keyser, E., Handa, T., & De Riek, J. (2007). Comparative study of the discriminating capacity and effectiveness of AFLP, STMS and EST markers in assessing genetic relationships among evergreen azales. Plant Breeding, 126, 207–212.
Singh, D. N. (2006). Participatory plant breeding as a method of rice breeding. International Rice Research Notes, 31(2), 48–50.
Sneath, P. H. A., & Sokal, R. R. (1973). Numerical taxonomy. San Francisco: Freeman.
Snoussi, H., Duval, M. F., Garcia-Lor, A., Belfalah, Z., Froelicher, Y., Risterucci, A. M., Perrier, X., Jacquemoud-Collet, J. P., Navarro, L., Harrabi, M., & Ollitrault, P. (2012). Assessment of the genetic diversity of the Tunisian citrus rootstock germplasm. BMC Genetics, 13(16), 1471–2156.
Sokal, R., & Michener, C. (1958). A statistical method for evaluating statistical relationships. Kansas University Science Bulletin, 38, 1409–1438.
Souframanien, J., & Gopalakrishna, T. (2004). A comparative analysis of genetic diversity in blackgram genotypes using RAPD and ISSR markers. Theoretical and Applied Genetics, 109, 1687–1693.
Sugawara, K., Wakizuka, T., Oowada, A., Moriguchi, T., & Omura, M. (2002). Histogenic identification by RAPD analysis of leaves and fruit of newly synthesized chimeric Citrus. Journal of the American Society for Horticultural Science, 127, 104–107.
Sun, G. L., William, M., Liu, J., Kasha, K. J., & Pauls, K. P. (2001). Microsatellite and RAPD polymosphisms in Ontario corn hybridos are related to the commercial sources and maturity ratings. Molecular Breeding, 7, 13–24.
Swingle, W. T., & Reece, P. C. (1967). The botany of Citrus and its wild relatives. In W. Reuther, L. D. Batchelor, & H. J. Webber (Eds.), The citrus industry (Vol. 1, pp. 190–430). Riverside: University of California.
Thomas, M. R., & Scott, N. S. (1993). Microsatellite repeats in grapevine reveal DNA polymorphism when analysed as sequence-tagged sites (STSs). Theoretical and Applied Genetics, 86, 985–990.
Tiwari Sh, K., Karihaloo, J. L., Hameed, N., & Gaikwad, A. B. (2009). Molecular characterization of brinjal (Solanum melongena L.) cultivars using RAPD and ISSR markers. Journal of Plant Biochemistry and Biotechnology, 18, 189–195.
Uzun, A., Yesiloglu, T., Aka-Kacar, Y., Tuzcu, O., & Gulsen, O. (2009). Genetic diversity and relationships within Citrus and related genera based on sequence related amplified polymorphism markers (SRAPs). Scientia Horticulturae, 121, 306–312.
Varshney, R. K., Chabane, K., Hendre, P. S., Aggarwal, R. K., & Graner, A. (2007). Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and Elite Barleys. Plant Science, 173, 638–649.
Wattier, R., Engel, C. R., Saumitou-Laprade, P., & Valero, M. (1998). Short allele dominance as a source of heterozygote deficiency at microsatellite loci: experimental evidence at the dinucleotide locus Gv1CT in Gracilaria gracilis (Rhodophyta). Molecular Ecology, 7, 1569–1573.
Weising, K., Nybom, H., Wolf, K., & Meyer, W. (1995). DNA fingerprinting in plants and fungi. Boca Rato: CRC Press.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A., & Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18, 6531–6535.
Acknowledgments
This work was supported by grants from the Tunisian “Ministry of Higher Education and Scientific Research”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lamine, M., Mliki, A. Elucidating Genetic Diversity among Sour Orange Rootstocks: a Comparative Study of the Efficiency of RAPD and SSR Markers. Appl Biochem Biotechnol 175, 2996–3013 (2015). https://doi.org/10.1007/s12010-015-1477-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1477-6