Abstract
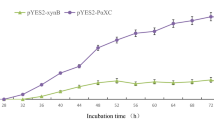
The aim of this study was to endow an industrial strain of Saccharomyces cerevisiae with the ability to overexpress the xylanase by constructing a homology-driven integration vector. The total mRNA from a xylanase-producing strain of Aspergillus niger IME-216 was extracted and used as the template for the production of endo-β-1,4-xylanase cDNA by reverse transcription. The fusion fragment containing the phosphoglycerate kinase promoter, α-factor signal peptide, xylanase gene encoding the mature peptide, and CYC1 terminator was first generated by overlap extension polymerase chain reaction. Then, the vector pUPX was constructed by inserting the fusion fragment into the S. cerevisiae plasmid pUG6. Then, A 2.2-kb rDNA sequence was further cloned and attached to the SalI-digested pUPX to obtain the integration plasmid pUPXR. The pUPXR was linearized by KpnI, transformed into the industrial strain S. cerevisiae YS2 using the lithium acetate method and integrated into the S. cerevisiae chromosome. The maximum yield of the recombinant xylanase produced by the engineered S. cerevisiae strain YS2_2 was 74.8 U per microliter, which was about 1.5-fold higher than the original 50 U per microliter by Aspergillus niger IME-216 strain under the flask culture at 28 °C for 72 h. The findings of our study can be used for further development of industrial S. cerevisiae strain for producing interested enzymes, or improving the achievement of metabolism, for example, simultaneous fermentation of glucose and xylose to producing bioethanol.



Similar content being viewed by others
References
Srinivasan, M. C., & Rele, M. V. (1999). Microbial xylanases for paper industry. Current Science, 77, 137–142.
Sunna, A., & Antranikian, G. (1996). Growth and production of xylanolytic enzymes by the extreme thermophilic anaerobic bacterium Thermotoga thermarum. Applied Microbiology and Biotechnology, 45, 671–676.
Whistler, R., & Richards, E. (1970). Hemicelluloses. In W. Pigman, D. Horton, & J. Wander (Eds.), The Carbohydrates (pp. 447–769). New York: Academic Press.
Biely, P. (1985). Microbial xylanolytic systems. Trends in Biotechnology, 3, 286–290.
Puls, J., Schmidt, O., & Granzow, C. (1987). Glucuronidase in two microbial xylanolytic systems. Enzyme and Microbial Technology, 9, 83–88.
Polizeli, M. L. T. M., Rizzatti, A. C. S., Monti, R., Terenzi, H. F., Jorge, J. A., & Amorim, D. S. (2005). Xylanases from fungi: properties and industrial applications. Applied Microbiology and Biotechnology, 67, 577–591.
Collins, T., Gerday, C., & Feller, G. (2005). Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiology Revview, 29, 3–23.
Twomey, L. N., Pluske, J. R., Rowe, J. B., Choct, M., Brown, W., McConnell, M. F., et al. (2003). The effects of increasing levels of soluble non-starch polysaccharides and inclusion of feed enzymes in dog diets on faecal quality and digestibility. Animal Feed Science and Technology, 108, 71–82.
Kim, J. H., Block, D. E., & Mills, D. A. (2010). Simultaneous consumption of pentose and hexose sugars: an optimal microbial phenotype for efficient fermentation of lignocellulosic biomass. Applied Microbiology and Biotechnology, 88, 1077–1085.
Jeffries, T. W., & Jin, Y.-S. (2004). Metabolic engineering for improved fermentation of pentoses by yeasts. Applied Microbiology and Biotechnology, 63, 495–509.
Bera, A. K., Ho, N. W., Khan, A., & Sedlak, M. (2010). A genetic overhaul of Saccharomyces cerevisiae 424A (LNH-ST) to improve xylose fermentation. Journal of Industrial Microbiology and Biotechnology, 38, 617–626.
Khramtsov, N., McDade, L., Amerik, A., Yu, E., Divatia, K., Tikhonov, A., et al. (2011). Industrial yeast strain engineered to ferment ethanol from lignocellulosic biomass. Bioresource Technology, 102, 8310–8313.
Madhavan, A., Srivastava, A., Kondo, A., & Bisarial, V. S. (2012). Bioconversion of lignocellulose-derived sugars to ethanol by engineered Saccharomyces cerevisiae. Critical Reviews in Biotechnology, 32, 22–48.
Xu, Y., Tian, B. Y., Jiang, X. Z., Huang, Q. G., Wang, C. X., & Huang, J. Z. (2009). Identification of a xylanase-producing fungi and its orthogonal experiment of fermentation conditions. Biotechnology (Chinese), 19, 51–55.
La Grange, D. C., Pretorius, I. S., Claeyssens, M., & Van Zyl, W. H. (2001). Degradation of xylan to D-xylose by recombinant Saccharomyces cerevisiae coexpressing the Aspergillus niger-xylosidase (xlnD) and the Trichoderma reesei xylanase II (xyn2) genes. Applied Environment Microbiology, 67, 5512–5519.
Murray, M. G., & Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8, 4321–4326.
Hill, J., Donald, K. A., & Griffiths, D. E. (1991). DMSO-enhanced whole cell yeast transformation. Nucleic Acids Research, 19, 5791.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press.
Pel, H. J., de Winde, J. H., Archer, D. B., Dyer, P. S., Hofmann, G., Schaap, P. J., et al. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology, 25, 221–231.
Biely, P., Vrsansk, M., Tenkanen, M., & Kluepfel, D. (1997). Endo-β-1,4-xylanase families: differences in catalytic properties. Journal of Biotechnology, 57, 151–166.
Henrissat, B., & Davies, G. (1997). Structural and sequence-based classification of glycoside hydrolases. Current Opinion in Structural Biology, 7, 637–644.
Ito, K., Ikemasu, T., & Ishikawa, T. (1992). Cloning and sequencing of the xynA gene encoding xylanase A of Aspergillus kawachii. Bioscience, Biotechnology, and Biochemistry, 56, 906–912.
Lopes, T. S., Wijs, I. J. D., Steenhauer, S. I., Verbakel, J., & Planta, R. J. (1996). Factors affecting the mitotic stability of high copy number integration into the ribosomal DNA of Saccharomyces cerevisiae. Yeast, 12, 467–477.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 31170108 and 30800735) and the earmarked fund for Modern Agro-industry Technology Research System (CARS-20-4-4).
Author information
Authors and Affiliations
Corresponding author
Additional information
B.Y. Tian and Y. Xu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Tian, B., Xu, Y., Cai, W. et al. Molecular Cloning and Overexpression of an Endo-β-1,4-xylanase Gene from Aspergillus niger in Industrial Saccharomyces cerevisiae YS2 Strain. Appl Biochem Biotechnol 170, 320–328 (2013). https://doi.org/10.1007/s12010-013-0173-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0173-7