Abstract
Purpose of Review
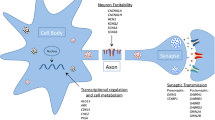
Seizures can arise in neocortical, thalamocortical, limbic or brainstem networks. Here, we review recent genetic mechanisms implicated in focal and genetic generalized epilepsies (GGEs).
Recent Findings
Pathogenic variation in GAP activity toward RAGs 1 (GATOR1) complex genes (i.e., DEPDC5, NPRL2 and NPRL3) mainly result in focal epilepsies. They are associated with high rates of sudden unexpected death in epilepsy and malformations of cortical development (MCD), where “two-hits” in GATOR1-related pathways are also found in MCDs. Large-scale sequencing studies continue to reveal new genetic risk (germline or somatic) variants, and new genes relevant to epileptic encephalopathies (EEs). Genes previously associated with EEs, including GABAA receptor genes, are now known to play a role in both common focal and GGEs in individuals without intellectual disabilities. These findings suggest that there may be a common pathophysiological mechanism in GGEs and focal epilepsies. Finally, polygenic risk scores, based on common genetic variation, offer promise in helping to differentiate between GGEs and common forms of focal epilepsies.
Summary
Genetic abnormalities are a significant cause of common sporadic epilepsies, epilepsies associated with inflammatory markers, and focal epilepsies with or without MCD. Future studies using genome sequencing may provide more answers to the remaining unresolved epilepsy cases.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Ngugi AK, Bottomley C, Kleinschmidt I, Sander JW, Newton CR. Estimation of the burden of active and life-time epilepsy: a meta-analytic approach. Epilepsia. 2010;51(5):883–90.
• Devinsky O, Vezzani A, O TJ, Jette N, Scheffer IE, de Curtis M, et al. Epilepsy. Nat Rev Dis Prim. 2018;3. This is a recent review of the epidemiology and pathophysiology of epilepsy.
Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512–21.
Fisher RS, Cross JH, French JA, Higurashi N, Hirsch E, Jansen FE, et al. Operational classification of seizure types by the International League Against Epilepsy: position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58(4):522–30.
Helbig I, Heinzen EL, Mefford HC, Berkovic SF, Lowenstein DH, Kato M, et al. Primer part I - the building blocks of epilepsy genetics. Epilepsia. 2016;57(6):861–8.
Oyrer J, Maljevic S, Scheffer IE, Berkovic SF, Petrou S, Reid CA. Ion channels in genetic epilepsy: from genes and mechanisms to disease-targeted therapies. Pharmacol Rev. 2018;70(1):142–73.
Myers CT, Mefford HC. Advancing epilepsy genetics in the genomic era. Genome Med. 2015;7(1):1–11.
Dunn P, Albury CL, Maksemous N, Benton MC, Sutherland HG, Smith RA, et al. Next generation sequencing methods for diagnosis of epilepsy syndromes. Front Genet. 2018;9(FEB):1–11.
•• Wang J, Lin ZJ, Liu L, Xu HQ, Shi YW, Yi YH, et al. Epilepsy-associated genes. Seizure. 2017;44:11–20 This study identifies 977 epilepsy-associated genes, and classifies them further into primary epilepsy genes, neurodevelopment associated epilepsy genes, epilepsy-related genes and potential epilepsy-associated genes.
Kurahashi H, Hirose S. Autosomal dominant nocturnal frontal lobe epilepsy. In: GeneReviews®. Seattle (WA): University of Washington, Seattle; 2018. p. 1–21.
Steinlein OK, Mulley JC, Propping P, Wallace RH, Phillips HA, Sutherland GR, et al. A missense mutation in the neuronal nicotinic acetylcholine receptor a-4 subunit is associated with autosomal dominant noctural frontal lobe epilepsy. Nat Genet. 1995;11:201–3.
Becchetti A, Aracri P, Meneghini S, Brusco S, Amadeo A. The role of nicotinic acetylcholine receptors in autosomal dominant nocturnal frontal lobe epilepsy. Front Physiol. 2015;6(February):1–12.
Klaassen A, Glykys J, Maguire J, Labarca C, Mody I, Boulter J. Seizures and enhanced cortical GABAergic inhibition in two mouse models of human autosomal dominant nocturnal frontal lobe epilepsy. Proc Natl Acad Sci U S A. 2006;103(50):19152–7.
Bertrand D. Neuronal nicotinic acetylcholine receptors and epilepsy. Epilepsy Curr. 2002;2(6):191–3.
Aridon P, Marini C, Di Resta C, Brilli E, De Fusco M, Politi F, et al. Increased sensitivity of the neuronal nicotinic receptor α2 subunit causes familial epilepsy with nocturnal wandering and ictal fear. Am J Hum Genet. 2006;79(2):342–50.
Hoda J, Gu W, Friedli M, Phillips HA, Bertrand S, Antonarakis SE, et al. Human nocturnal frontal lobe epilepsy : Pharmocogenomic profiles of pathogenic nicotinic acetylcholine receptor. Mol Pharmacol. 2008;74(2):379–91.
Scheffer IE, Bhatia KP, Lopes-cendes I, Fish DR, Marsden CD, Andermann E, et al. Autosomal dominant nocturnal frontal lobe epilepsy: a distinctive clinical disorder. Brain. 1995;118(1):61–73.
De Fusco M, Becchetti A, Patrignani A, Annesi G, Gambardella A, Quattrone A, et al. The nicotinic receptor β2 subunit is mutant in nocturnal frontal lobe epilepsy. Nat Genet. 2000;26:275–6.
Phillips HA, Favre I, Kirkpatrick M, Zuberi SM, Goudie D, Heron SE, et al. CHRNB2 is the second acetylcholine receptor subunit associated with autosomal dominant nocturnal frontal lobe epilepsy. Am J Hum Genet. 2001;68(1):225–31.
Heron SE, Smith KR, Bahlo M, Nobili L, Kahana E, Licchetta L, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 2012;44(11):1188–90.
Dibbens LM, De Vries B, Donatello S, Heron SE, Hodgson BL, Chintawar S, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet. 2013;45(5):546–51.
Ostendorf AP, Wong M. mTOR inhibition in epilepsy: rationale and clinical perspectives. CNS Drugs. 2015;29(2):91–9.
Ricos MG, Hodgson BL, Pippucci T, Saidin A, Ong YS, Heron SE, et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol. 2016;79(1):120–31.
Nascimento FA, Borlot F, Cossette P, Minassian BA, Andrade DM. Two definite cases of sudden unexpected death in epilepsy in a family with a DEPDC5 mutation. Neurol Genet. 2015;1:e28.
• Baldassari S, Picard F, Verbeek NE, van Kempen M, Brilstra EH, Lesca G, et al. The landscape of epilepsy-related GATOR1 variants. Genet Med. 2019;21(2):398–408 This study reported clinical and molecular genetic data in a total of 73 individivuals with rare pathogenic variation impacting the GATOR1 complex genes (DEPDC5, NPRL2 or NPRL3), providing a review of the landscape of epilepsy-related GATOR1 variants.
Martin C, Meloche C, Rioux MF, Nguyen DK, Carmant L, Andermann E, et al. A recurrent mutation in DEPDC5 predisposes to focal epilepsies in the French-Canadian population. Clin Genet. 2014;86(6):570–4.
Ishida S, Picard F, Rudolf G, Noé E, Achaz G, Thomas P, Genton P, Mundwiller E, Wolff M, Marescaux C, Miles R, Baulac M, Hirsch E, Leguern E, Baulac S Mutations of DEPDC5 cause autosomal dominant focal epilepsies. Nat Genet 2013;45(5):552–5, 555.
Picard F, Makrythanasis P, Navarro V, Ishida S, De Bellescize J, Ville D, et al. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology. 2014;82(23):2101–6.
Scheffer IE, Heron SE, Regan BM, Mandelstam S, Crompton DE, Hodgson BL, et al. Mutations in mammalian target of rapamycin regulator DEPDC5 cause focal epilepsy with brain malformations. Ann Neurol. 2014;75(5):782–7.
Bagnall RD, Crompton DE, Petrovski S, Lam L, Cutmore C, Garry SI, et al. Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol. 2016;79(4):522–34.
Lasarge CL, Danzer SC. Mechanisms regulating neuronal excitability and seizure development following mTOR pathway hyperactivation. Front Mol Neurosci. 2014;7(March):1–15.
Harden C, Tomson T, Gloss D, Buchhalter J, Cross JH, Donner E, et al. Practice guideline summary: sudden unexpected death in epilepsy incidence rates and risk factors. Neurology. 2017;88(17):1674–80.
Devinsky O, Hesdorffer DC, Thurman DJ, Lhatoo S, Richerson G. Sudden unexpected death in epilepsy: epidemiology, mechanisms, and prevention. Lancet Neurol. 2016;15(10):1075–88.
Baulac S, Ishida S, Marsan E, Nordli D, Cossette P, Nguyen S, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol. 2015;77:675–83.
D’Gama AM, Geng Y, Couto J, Martin B, Boyle E, LaCoursiere C, et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol. 2015;77:720–5.
Sim JC, Scerri T, Fanjul-fern M, Riseley JR, Gillies G, Pope K, et al. Familial cortical dysplasia caused by mutation in the mammalian target of rapamycin regulator NPRL3. Ann Neurol. 2016;79:132–7.
Weckhuysen S, Marsan E, Lambrecq V, An-gourfinkel I, Baulac M, Fohlen M. Involvement of GATOR complex genes in familial focal epilepsies and focal cortical dysplasia. Epilepsia. 2016;57(6):994–1003.
Represa A. Why malformations of cortical development cause epilepsy. Front Neurosci. 2019;13(March):1–10.
Blümcke I, Thom M, Aronica E, Armstrong DD, Vinters HV, Palmini A, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52(1):158–74.
Blumcke I, Spreafico R, Haaker G, Coras R, Kobow K, Bien CG, et al. Histopathological findings in brain tissue obtained during epilepsy surgery. N Engl J Med. 2017;377(17):1648–56.
• Lim JS, Kim WI, Kang HC, Kim SH, Park AH, Park EK, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21(4):395–400. This is one of the earliest studies to provide evidence that brain somatic variation in MTOR can cause focal cortical dysplasia.
Ribierre T, Deleuze C, Bacq A, Baldassari S, Marsan E, Chipaux M, et al. Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J Clin Invest. 2018;128(6):2452–8.
D’Gama AM, Woodworth MB, Hossain AA, Bizzotto S, Hatem NE, LaCoursiere CM, et al. Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 2017;21(13):3754–66.
Uddin M, Woodbury-Smith M, Chan A, Brunga L, Lamoureux S, Pellecchia G, et al. Germline and somatic mutations in STXBP1 with diverse neurodevelopmental phenotypes. Neurol Genet. 2017;3(6).
Winawer MR, Griffin NG, Samanamud J, Baugh EH, Rathakrishnan D, Ramalingam S, et al. Somatic SLC35A2 variants in the brain are associated with intractable neocortical epilepsy. Ann Neurol. 2018;83(6):1133–46.
Koh HY, Lee JH. Brain somatic mutations in epileptic disorders. Mol Cells. 2018;41(10):881–8.
D’Gama AM, Walsh CA. Somatic mosaicism and neurodevelopmental disease. Nat Neurosci. 2018;21(11):1504–14.
Blümcke I, Thom M, Wiestler OD. Ammon’s horn sclerosis: a maldevelopmental disorder associated with temporal lobe epilepsy. Brain Pathol. 2006;12(2):199–211.
Leverenz JB, Agustin CM, Tsuang D, Peskind ER, Edland SD, Nochlin D, et al. Clinical and neuropathological characteristics of hippocampal sclerosis: a community-based study. Arch Neurol. 2002;59(7):1099–106.
Wiencken-Barger AE, Djukic B, Casper KB, McCarthy KD. A role for Connexin43 during neurodevelopment. Glia. 2007;55:675–86.
Collignon F, Wetjen NM, Cohen-Gadol AA, Cascino GD, Parisi J, Meyer FB, et al. Altered expression of connexin subtypes in mesial temporal lobe epilepsy in humans. J Neurosurg. 2006;105(1):77–87.
•• The International League Against Epilepsy Consortium on Complex Epilepsies. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. 2018;9:5269 This study reports the largest genome-wide mega-analysis to date, identifying a total of 16 loci using 15,212 individuals with epilepsy and 29,677 controls.
Johnson MR, Behmoaras J, Bottolo L, Krishnan ML, Pernhorst K, Santoscoy PLM, et al. Systems genetics identifies Sestrin 3 as a regulator of a proconvulsant gene network in human epileptic hippocampus. Nat Commun. 2015;6.
Huang LG, Zou J, Lu QC. Silencing rno-miR-155-5p in rat temporal lobe epilepsy model reduces pathophysiological features and cell apoptosis by activating Sestrin-3. Brain Res. 1689;2018:109–22.
Henshall DC, Hamer HM, Pasterkamp RJ, Goldstein DB, Kjems J, Prehn JHM, et al. MicroRNAs in epilepsy: pathophysiology and clinical utility. Lancet Neurol. 2016;15(13):1368–76. https://doi.org/10.1016/S1474-4422(16)30246-0.
Li TR, Jia YJ, Wang Q, Shao XQ, Zhang P, Lv RJ. Correlation between tumor necrosis factor alpha mRNA and microRNA-155 expression in rat models and patients with temporal lobe epilepsy. Brain Res. 1700;2018:56–65.
Mullen SA, Berkovic SF, Berkovic SF, Lowenstein DH, Kato M, Cross H, et al. Genetic generalized epilepsies. Epilepsia. 2018;59(6):1148–53.
Berkovic SF, Howell RA, Hay DA, Hopper JL. Epilepsies in twins: genetics of the major epilepsy syndromes. Ann Neurol. 1998;43(4):435–45.
Lachance-Touchette P, Brown P, Meloche C, Kinirons P, Lapointe L, Lacasse H, et al. Novel α1 and γ2 GABA a receptor subunit mutations in families with idiopathic generalized epilepsy. Eur J Neurosci. 2011;34(2):237–49.
Kang J, Macdonald RL. The GABAA receptor γ2 subunit R43Q mutation linked to childhood absence epilepsy and febrile seizures causes retention of α1β2α2S receptors in the endoplasmic reticulum. J Neurosci. 2004;24(40):8672–7.
Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, et al. 15Q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41(2):160–2.
• De Kovel CGF, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133(1):23–32 This study investigated the impact of five microdeletions (1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2) on the genetic risk to common idiopathic GGEs.
Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148(6):1223–41.
Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, et al. Exome sequencing of ion channel genes reveals complex variant profiles confounding personal risk assessment in epilepsy. Cell. 2012;145(7):1036–48.
•• Heinzen EL, Depondt C, Cavalleri GL, Ruzzo EK, Walley NM, Need AC, et al. Exome sequencing followed by large-scale genotyping fails to identify single rare variants of large effect in idiopathic generalized epilepsy. Am J Hum Genet. 2012;91(2):293–302 This study examined whole-exome sequencing data from 118 individuals with idiopathic generalized epilepsy, finding no variants with statistically significant associations.
•• Allen AS, Bellows ST, Berkovic SF, Bridgers J, Burgess R, Cavalleri G, et al. Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017;16(2):135–43 In this whole exome sequencing study, the contribution of ultra-rare genetic variation was assessed in common epilepsies, finding excess ultra-rare variation in known epilepsy genes.
•• May P, Girard S, Harrer M, Bobbili DR, Schubert J, Wolking S, et al. Rare coding variants in genes encoding GABA-A receptors in genetic generalised epilepsies: an exome-based case-control study. Lancet Neurol. 2018;17(8):699–708 This whole-exome based case-control study investigated the burden of rare genetic variants in GGEs, finding that functionally relevant GABAA variants are a significant genetic risk factor for GGEs.
Shen D, Hernandez CC, Shen W, Hu N, Poduri A, Shiedley B, et al. De novo GABRG2 mutations associated with epileptic encephalopathies. Brain. 2017;140(1):49–67.
Johannessen K, Marini C, Pfeffer S, Møller RS, Dorn T, Niturad C, et al. Phenotypic spectrum of GABRA1: from generalized epilepsies to severe epileptic encephalopathies. Neurology. 2016;87(11):1140–51.
Møller RS, Wuttke TV, Helbig I, Marini C, Johannesen KM, Brilstra EH, et al. Mutations in GABRB3: from febrile seizures to epileptic encephalopathies. Neurology. 2017;88(5):483–92.
Wolking S, May P, Mei D, Møller RS, Balestrini S, Helbig KL, et al. Clinical spectrum of STX1B -related epileptic disorders. Neurology. 2019;92(11):E1238–49.
Koepp MJ, Thomas RH, Wandschneider B, Berkovic SF, Schmidt D. Concepts and controversies of juvenile myoclonic epilepsy: still an enigmatic epilepsy. Expert Rev Neurother. 2014;14(7):819–31.
Curwood EK, Pedersen M, Carney PW, Berg AT, Abbott DF, Jackson GD. Abnormal cortical thickness connectivity persists in childhood absence epilepsy. Ann Clin Transl Neurol. 2015;2(5):456–64.
Chowdhury FA, Elwes RDC, Koutroumanidis M, Morris RG, Nashef L, Richardson MP. Impaired cognitive function in idiopathic generalized epilepsy and unaffected family members: An epilepsy endophenotype. Epilepsia. 2014;55(6):835–40.
Holmes MD, Brown M, Tucker DM. Are “generalized” seizures truly generalized? Evidence of localized mesial frontal and frontopolar discharges in absence. Epilepsia. 2004;45(12):1568–79.
Sugrue LP, Desikan RS. What are polygenic scores and why are they important? JAMA - J Am Med Assoc. 2019;321(18):1820–1.
•• Leu C, Stevelink R, Smith AW, Goleva SB, Kanai M, Ferguson L, et al. Polygenic burden in focal and generalized epilepsies. Brain. 2019;142(11):3473–81 This study calculated polygenic risk scores for GGE and focal epilepsies using data from the International League Against Epilepsy Consortium on Complex Epilepsies (2018).
Ishiura H, Doi K, Mitsui J, Yoshimura J, Matsukawa MK, Fujiyama A, et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet. 2018;50(4):581–90.
Yeetong P, Pongpanich M, Srichomthong C, Assawapitaksakul A, Shotelersuk V, Tantirukdham N, et al. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain. 2019;142:3360–6.
Corbett MA. Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. (2019):1–61.
Florian RT, Kraft F, Leitão E, Kaya S, Klebe S, Magnin E, et al. Unstable TTTTA/TTTCA expansions in MARCH6 are associated with familial adult myoclonic epilepsy type 3. Nat Commun. 2019;10:4919.
Cen Z, Jiang Z, Chen Y, Zheng X, Xie F, Yang X, et al. Intronic pentanucleotide TTTCA repeat insertion in the SAMD12 gene causes familial cortical myoclonic tremor with epilepsy type 1. Brain. 2018;141(8):2280–8.
Ostrander BEP, Butterfield RJ, Pedersen BS, Farrell AJ, Layer RM, Ward A, et al. Whole-genome analysis for effective clinical diagnosis and gene discovery in early infantile epileptic encephalopathy. npj Genomic Med. 2018;3(1):22.
Martin HC, Kim GE, Pagnamenta AT, Murakami Y, Carvill GL, Meyer E, et al. Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum Mol Genet. 2014;23(12):3200–11.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
DMA has received research grants from the McLaughlin Centre, EpLink and the Dravet Syndrome Foundation. DMA is a part of the MAB of Stoke Therapeutics and has received consulting fees from Eisai.
Human and Animal Rights and Informed Consent
All reported studies/experiments with human or animal subjects performed by the authors have been previously published (unless stated otherwise) and compiled with all applicable ethical standards (including the Helsinki declaration and its amendments, institutional/national research committee standards, and international/national/institutional guidelines).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Epilepsy
Rights and permissions
About this article
Cite this article
Qaiser, F., Yuen, R.K.C. & Andrade, D.M. Genetics of Epileptic Networks: from Focal to Generalized Genetic Epilepsies. Curr Neurol Neurosci Rep 20, 46 (2020). https://doi.org/10.1007/s11910-020-01059-x
Published:
DOI: https://doi.org/10.1007/s11910-020-01059-x