Abstract
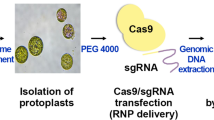
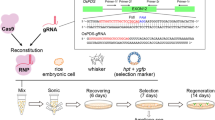
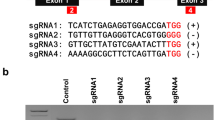
Nowadays, genome editing in plants has become much easier thanks to the recently developed clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 9 (CRISPR–Cas9) nuclease system. However, to combine protoplast technology with the CRISPR–Cas9 system in plants, a stable and an efficient foreign DNA delivery system is essential for gene editing. In the present study, we developed an electro-transfection system for CRISPR–Cas9 ribonucleoprotein (RNP) delivery to cabbage protoplasts. Under 1000 V treatment, the frequency of initial cell division and total number of cell colonies formed were 47.7 ± 2.5% and 52 ± 7.5%, respectively. The total number of cell colonies formed following 1000 V treatment was 1.4 times higher than that following polyethylene glycol (PEG) treatment. However, the frequency of initial cell division and total number of cell colonies formed from protoplasts decreased with increasing voltage. Cy3–Cas9 protein delivery into the nucleus was confirmed through both electro-transfection and PEG-mediated transfection using confocal laser scanning microscopy. The frequency of insertions and deletions in the synthesized guide RNA of phytoene desaturase 1 was the highest at 3.4% following electro-transfection at 1000 V with a pulse width of 20 ms and only 1.8% following PEG-mediated transfection. These results indicate that electro-transfection is more efficient in RNP delivery to protoplast than PEG-mediated transfection in cabbage for PDS1 sgRNA delivery. Therefore, the electro-transfection system developed in the present study presents the possibility it could be used for DNA-free genome editing of other crops.



Similar content being viewed by others
References
Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, Kemaux PG, Medford JI, Orozco-Cardenas ML, Tricoli DM, Van Eck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Neal Stewark C (2016) Advancing crop transformation in the era of genome editing. Plant Cell 28:1510–1520
Andersson M, Turesson H, Olsson N, Fält A-S, Ohlsson P, Gonzalez MN, Samuelsson M, Hofvander P (2018) Genome editing in potato via CRISPR–Cas9 ribonucleoprotein delivery. Physiol Plant 164(4):378–384
Bhowmik P, Ellison E, Polley B, Bollina V, Kulkarmi M, Ghanbarnia K, Song H, Gao C, Voytas DF, Kagale S (2018) Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Scientific reports 8:6502
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Brees C, Fransen M (2014) A cost-effective approach to microporate mammalian cells with the neon transfection system. Anal Biochem 466:49–50
De Buck S, De Wilde C, Van Montagu M, Depicker A (2000) T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol Breeding 6:459–468
Deng W, Shi X, Tjian R, Lionnet T, Singer RH (2015) CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci 112:11870–11875
Frearson EM, Power JB, Cocking EC (1973) The isolation, culture and regeneration of Petunia leaf protoplasts. Dev Biol 33:130–137
Gelvin SB (2017) Integration of Agrobacterium T-DNA into the plant genome. Annu Rev Gene 51:195–217
Jie EY, Kim SW, Jang HR, In DS, Liu JR (2011) Myo-inositol increases the plating efficiency of protoplast derived from cotyledon of cabbage (Brassica oleracea var. capitata). J Plant Biotechnol 38:69–76
Jupe F, Rivkin AC, Michael TP, Zander M, Motley ST, Sandoval JP, Slotkin RK, Chen H, Castanon R, Nery JR, Ecker JR (2019) The complex architecture and epigenomic impact of plant T-DNA insertions. PLOS Genet 15:e1007819
Kao KN, Michayluk MR (1974) A method for high-frequency intergeneric fusion of plant protoplasts. Planta 115:355–367
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8:14261
Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Nagamangala Kanchiswamy C (2016) DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci 7:1904
Metje-Sprink J, Menz J, Modrzejewski D, Sprink T (2019) DNA-free genome editing: past, present and future. Front Plant Sci 9:1957
Murovec J, Guček K, Bohanec B, Avbelj M, Jerala R (2018) DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR–Cas9 ribonucleoprotein complexes. Front Plant Sci 9:1594
Park S-C, Park S, Jeong YJ, Lee SB, Pyun JW, Kim S, Kim TH, Kim SW, Jeong JC, Kim CH (2019) DNA-free mutagenesis of GIGANTEA in Brassica oleracea var. capitata using CRISPR/Cas9 ribonucleoprotein complexes. Plant Biotechnol Rep 13:483–489
Sanford JC, Klein TM, Wolf ED, Allen N (1987) Delivery of substances into cells and tissues using a particle bombardment process. Particul Sci Technol 5:27–37
Schumann K, Lin S, Boyer E, Simeonow DDR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, Marson A (2015) Generation of knock-in primary human T cell using Cas9 ribonucleoproteins. Proc Natl Acad Sci 112:10437–10442
Shou H, Frame BR, Whitham SA, Wang K (2004) Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breeding 13:201–208
Song G, Jia K, Chen K, Kong X, Khattak B, Xie C, Li A, Mao L (2016) CRISPR/Cas9: a powerful tool for crop genome editing. Crop J 4:75–82
Subburaj S, Chung SJ, Lee C, Ryu S-M, Kim DH, Kim J-S, Bae S, Lee G-J (2016) Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep 35:1535–1544
Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nature Commun 7:13274
Tyagi S, Spörlein B, Tyagi AK, Herrmann RG, Koop HU (1989) PEG-and electroporation-induced transformation in Nicotiana tabacum: influence of genotype on transformation frequencies. Theor Appl Genet 78:287–292
Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim S-G, Kim S-T, Choe S, Kim J-S (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Acknowledgements
This work was supported by a grant from the KRIBB Research Initiative Program (KGM5282021); WISET-2020-193; a grant from the New breeding technologies development Program (Project No. PJ01480201), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, M.H., Lee, J., Choi, S.A. et al. Efficient genome editing using CRISPR–Cas9 RNP delivery into cabbage protoplasts via electro-transfection. Plant Biotechnol Rep 14, 695–702 (2020). https://doi.org/10.1007/s11816-020-00645-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-020-00645-2