Abstract
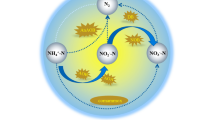
To improve nitrogen removal performance of wastewater treatment plants (WWTPs), it is essential to understand the behavior of nitrogen cycling communities, which comprise various microorganisms. This study characterized the quantity and diversity of nitrogen cycling genes in various processes of municipal WWTPs by employing two molecular-based methods:most probable number-polymerase chain reaction (MPN-PCR) and DNA microarray. MPN-PCR analysis revealed that gene quantities were not statistically different among processes, suggesting that conventional activated sludge processes (CAS) are similar to nitrogen removal processes in their ability to retain an adequate population of nitrogen cycling microorganisms. Furthermore, most processes in the WWTPs that were researched shared a pattern:the nirS and the bacterial amoA genes were more abundant than the nirK and archaeal amoA genes, respectively. DNA microarray analysis revealed that several kinds of nitrification and denitrification genes were detected in both CAS and anaerobic-oxic processes (AO), whereas limited genes were detected in nitrogen removal processes. Results of this study suggest that CAS maintains a diverse community of nitrogen cycling microorganisms; moreover, the microbial communities in nitrogen removal processes may be specific.

Similar content being viewed by others
References
Smith V H, Tilman G D, Nekola J C. Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environmental Pollution, 1999, 100(1-3): 179–196
Ahn Y H. Sustainable nitrogen elimination biotechnologies: a review. Process Biochemistry, 2006, 41(8): 1709–1721
Richardson D J, Watmough N J. Inorganic nitrogen metabolism in bacteria. Current Opinion in Chemical Biology, 1999, 3(2): 207–219
Zhu G B, Peng Y Z, Li B K, Guo J H, Yang Q, Wang S Y. Biological removal of nitrogen from wastewater. Reviews of Environmental Contamination and Toxicology, 2008, 192: 159–195
Song B, Tobias C R. Molecular and stable isotope methods to detect and measure anaerobic ammonium oxidation (anammox) in aquatic ecosystems. Methods in Enzymology, 2011, 496: 63–89
Kampschreur M J, Temmink H, Kleerebezem R, Jetten M S M, van Loosdrecht C M. Nitrous oxide emission during wastewater treatment. Water Research, 2009, 43(17): 4093–4103
Soda S, Arai T, Inoue D, Ishigaki T, Ike M, Yamada M. Statistical analysis of global warming potential, eutrophication potential, and sludge production of wastewater treatment in Japan. Journal of Sustainable Energy and Environment, 2013, 4(1): 33–40
Amann R I, Ludwing W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews, 1995, 59(1): 143–169
Prosser J I, Embley T M. Cultivation-based and molecular approaches to characterization of terrestrial and aquatic nitrifiers. Antonie van Leeuwenhoek, 2002, 81(1): 165–179
Smith C J, Osborn A M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiology Ecology, 2009, 67(1): 6–20
He Z, Deng Y, Zhou J. Development of functional gene microarrays for microbial community analysis. Current Opinion in Biotechnology, 2012, 23(1): 49–55
Inoue D, Pang J, Matsuda M, Sei K, Nishida K, Ike M. Development of a whole community genome amplification-assisted DNA microarray method to detect functional genes involved in the nitrogen cycle. World Journal of Microbiology & Biotechnology, 2014, 30(11): 2907–2915
Hashimoto K, Matsuda M, Inoue D, Ike M. Bacterial community dynamics in a full-scale municipal wastewater treatment plant employing conventional activated sludge process. Journal of Bioscience and Bioengineering, 2014, 118(1): 64–71
Iwamoto T, Tani K, Nakamura K, Suzuki Y, Kitagawa M, Eguchi M, Nasu M. Monitoring impact of in situ biostimulation treatment on groundwater bacterial community by DGGE. FEMS Microbiology Ecology, 2000, 32(2): 129–141
Rotthauwe J H, de Boe W, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Applied and Environmental Microbiology, 1997, 63(12): 4704–4712
Sang N, Soda S, Sei K, Ike M. Effect of aeration on stabilization of organic solid waste and microbial population dynamics in lab-scale landfill bioreactors. Journal of Bioscience and Bioengineering, 2008, 106(5): 425–432
Francis C A, Roberts K J, Michael J, Santoro A E, Oakley B B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102 (41): 16683–14688
Wu L, Thompson D K, Li G, Hurt R A, Tiedje J M, Zhou J. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Applied and Environmental Microbiology, 2001, 67(12): 5780–5790
Wu L, Liu X, Christopher W S, Zhou J. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole community genome amplification. Applied and Environmental Microbiology, 2006, 72(7): 4931–4941
Geets J, de Cooman M, Wittebolle L, Heylen K, Vanparys B, de Vos P, Verstraete W, Boon N. Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Applied Microbiology and Biotechnology, 2007, 75(1): 211–221
Wang Z, Zhang X X, Lu X, Liu B, Li Y, Long C, Li A. Abundance and diversity of bacterial nitrifiers and denitrifiers and their functional genes in tannery wastewater treatment plants revealed by high-throughput sequencing. PLoS ONE, 2014, 9(11): e113603
Shu D, He Y, Yue H, Wang Q. Microbial structures and community functions of anaerobic sludge in six full-scale wastewater treatment plants as revealed by 454 high-throughput pyrosequencing. Bioresource Technology, 2015, 186: 163–172
Daims H, Wagner M. The microbiology of nitrogen removal. In: Seviour RJ, Nielsen PH, eds. Microbial Ecology of Activated Sludge. London: IWA Publishing, 2010,259–280
Kayee P, Sonthiphand P, Rongsayamanont C, Limpiyakorn T. Archaeal amoA genes outnumber bacterial amoA genes in municipal wastewater treatment plants in Bangkok. Microbial Ecology, 2011, 62(4): 776–788
Limpiyakorn T, Sonthiphand P, Rongsayamanont C, Polprasert C. Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresource Technology, 2011, 102(4): 3694–3701
Wu Y J, Whang L M, Fukushima T, Chang S H. Responses of ammonia-oxidizing archaeal and betaproteobacterial populations to wastewater salinity in a full-scale municipal wastewater treatment plant. Journal of Bioscience and Bioengineering, 2013, 115(4): 424–432
Burgin A J, Hamilton S K. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Frontiers in Ecology and the Environment, 2007, 5(2): 89–96
Kraft B, Strous M, Tegetmeyer H E. Microbial nitrate respiration— genes, enzymes and environmental distribution. Journal of Biotechnology, 2011, 155(1): 104–117
Rütting T, Boeckx P, Müller C, Klemedtsson L. Assessment of the importance of dissimilatory nitrate reduction to ammonium for the terrestrial nitrogen cycle. Biogeosciences, 2011, 8(7): 1779–1791
van den Berg E M, van Dongen U, Abbas B, van Loosdrecht M C M. Enrichment of DNRA bacteria in a continuous culture. ISME Journal, 2015, 9(10): 2153–2161
Strous M, van Gerven E, Kuenen J G, Jetten M. Effects of aerobic and microaerobic conditions on anaerobic ammonium-oxidizing (anammox) sludge. Applied and Environmental Microbiology, 1997, 63(6): 2446–2448
Matsuda M, Inoue D, Anami Y, Tsutsui H, Sei K, Soda S, Ike M. Comparative analysis of DNA-based microbial community composition and substrate utilisation patterns of activated sludge microorganisms from wastewater treatment plants operated under different conditions. Water Science and Technology, 2010, 61(11): 2843–2851
Takada K, Hashimoto K, Soda S, Ike M, Yamashita K, Hashimoto T. Characterization of microbial community in membrane bioreactors treating domestic wastewater. Journal of Water and Environment Technology, 2014, 12(2): 99–107
Ma Q, Qu Y, Shen W, Zhang Z, Wang J, Liu Z, Li D, Li H, Zhou J. Bacterial community compositions of coking wastewater treatment plants in steel industry revealed by Illumina high-throughput sequencing. Bioresource Technology, 2015, 179: 436–443
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Pang, J., Matsuda, M., Kuroda, M. et al. Characterization of the genes involved in nitrogen cycling in wastewater treatment plants using DNA microarray and most probable number-PCR. Front. Environ. Sci. Eng. 10, 7 (2016). https://doi.org/10.1007/s11783-016-0846-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11783-016-0846-x