Abstract
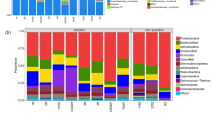
A rhizobox system constructed with crude oil-contaminated soil was vegetated with alfalfa (Medicago sativa L.) to evaluate the rhizosphere effects on the soil microbial population and functional structure, and to explore the potential mechanisms by which plants enhance the removal of crude oil in soil. During the 80-day experiment, 31.6% of oil was removed from the adjacent rhizosphere (AR); this value was 27% and 53%higher than the percentage of oil removed from the far rhizosphere (FR) and from the non-rhizosphere (NR), respectively. The populations of heterotrophic bacteria and hydrocarbon-degrading bacteria were higher in the AR and FR than in the NR. However, the removal rate of crude oil was positively correlated with the proportion of hydrocarbon-degrading bacteria in the rhizosphere. In total, 796, 731, and 379 functional genes were detected by microarray in the AR, FR, and NR, respectively. Higher proportions of functional genes related to carbon degradation and organic remediation, were found in rhizosphere soil compared with NR soil, suggesting that the rhizosphere selectively increased the abundance of these specific functional genes. The increase in water-holding capacity and decrease in pH as well as salinity of the soil all followed the order of AR>FR>NR. Canonical component analysis showed that salinity was the most important environmental factor influencing the microbial functional structure in the rhizosphere and that salinity was negatively correlated with the abundance of carbon and organic degradation genes.
Similar content being viewed by others
References
Liang Y, Zhang X, Dai D, Li G. Porous biocarrier-enhanced biodegradation of crude oil contaminated soil. International Biodeterioration & Biodegradation, 2009, 63(1): 80–87
Jia J, Li G, Zhong Y. Microbial eco-system of the contaminated soil with petrochemical hydrocarbons in the oilfields of northern China. In: Proceedings of the International Symposium on Water Resources and the Urban Environment 2003. Beijing: China Environmental Science Press, 216–220
van Agteren M H, Keuning S, Janssen D B. Handbook on Biodegradation and Biological Treatment of Hazardous Organic Compounds.Dordrecht: Kluwer Academic Publishers, 1999
Zhu L, Lu L, Zhang D. Mitigation and remediation technologies for organic contaminated soils. Frontiers of Environmental Science & Engineering in China, 2010, 4(4): 373–386
Susarla S, Medina V F, McCutcheon S C. Phytoremediation: an ecological solution to organic chemical contamination. Ecological Engineering, 2002, 18(5): 647–658
Wang Z, Xu Y, Zhao J, Li F, Gao D, Xing B. Remediation of petroleum contaminated soils through composting and rhizosphere degradation. Journal of Hazardous Materials, 2011, 190(1–3): 677–685
Gurska J, Wang W, Gerhardt K E, Khalid A M, Isherwood D M, Huang X D, Glick B R, Greenberg B M. Three year field test of a plant growth promoting rhizobacteria enhanced phytoremediation system at a land farm for treatment of hydrocarbon waste. Environmental Science & Technology, 2009, 43(12): 4472–4479
Peng S, Zhou Q, Cai Z, Zhang Z. Phytoremediation of petroleum contaminated soils by Mirabilis Jalapa L. in a greenhouse plot experiment. Journal of Hazardous Materials, 2009, 168(2–3): 1490–1496
Korade D L, Fulekar M H. Rhizosphere remediation of chlorpyrifos in mycorrhizospheric soil using ryegrass. Journal of Hazardous Materials, 2009, 172(2–3): 1344–1350
Lombi E, Wenzel W W, Gobran G R, Adriano D C. Dependency of phytoavailability of metals on indigenous and induced rhizosphere processes: a review. In: Gobran G R, Wenzel W W, Lombi E, eds. Trace Elements in the Rhizosphere. Boca Raton: CRC Press, 2001, 3–24
White PM, Wolf D C, Thoma G J, Reynolds CM. Phytoremediation of alkylated polycyclic aromatic hydrocarbons in a crude oil-contaminated soil. Water, Air, and Soil Pollution, 2006, 169(1–4): 207–220
Su Y, Yang X, Chiou C. Effect of rhizosphere on soil microbial community and in-situ pyrene biodegradation. Frontiers of Environmental Science & Engineering in China, 2008, 2(4): 468–474
Beauregard M S, Hamel C, Atul-Nayyar, St-Arnaud M. Long-term phosphorus fertilization impacts soil fungal and bacterial diversity but not AM fungal community in alfalfa. Microbial Ecology, 2010, 59(2): 379–389
Kirk J L, Klironomos J N, Lee H, Trevors J T. The effects of perennial ryegrass and alfalfa on microbial abundance and diversity in petroleum contaminated soil. Environmental Pollution, 2005, 133 (3): 455–465
Wenzel W W, Wieshammer G, Fitz W J, Puschenreiter M. Novel rhizobox design to assess rhizosphere characteristics at high spatial resolution. Plant and Soil, 2001, 237(1): 37–45
Norvell W A, Cary E E. Potential errors caused by roots in analyses of rhizosphere soil. Plant and Soil, 1992, 143(2): 223–231
Awad F, Römheld V, Marschner H. Effect of root exudates on mobilization in the rhizosphere and uptake of iron by wheat plants. Plant and Soil, 1994, 165(2): 213–218
Gahoonia T S, Nielsen N E. A method to study zhizosphere processes in thin soil layers of different proximity to roots. Plant and Soil, 1991, 135(1): 143–146 doi:10.1007/BF00014787
Gahoonia T S, Nielsen N E. Control of pH at the soil-root interface. Plant and Soil, 1992, 140(1): 49–54
Youssef R A, Chino M. Root induced changes in the rhizosphere of plants. I. Changes in relation to the bulk soil. Soil Science and Plant Nutrition, 1989, 35(3): 461–468
Youssef R A, Chino M. Root-induced changes in the rhizosphere of plants. II. Distribution of heavy metals across the rhizosphere in soils. Soil Science and Plant Nutrition, 1989, 35(4): 609–621
Kirk G J. A model of phosphate solubilization by organic anion excretion from plant roots. European Journal of Soil Science, 1999, 50(3): 369–378
He Z, Zhou J. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Applied and Environmental Microbiology, 2008, 74(10): 2957–2966
He Z L, van Nostrand J D, Wu L Y, Zhou J Z. Development and application of functional gene arrays for microbial community analysis. Transactions of Nonferrous Metals Society of China, 2008, 18(6): 1319–1327
He Z, Gentry T J, Schadt C W, Wu L, Liebich J, Chong S C, Huang Z, Wu W, Gu B, Jardine P, Criddle C, Zhou J. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. The ISME Journal, 2007, 1(1): 67–77
He Z, van Nostrand J D, Deng Y, Zhou J. Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Frontiers of Environmental Science & Engineering in China, 2011, 5(1): 1–20
Liang Y, van Nostrand J D, Wang J, Zhang X, Zhou J, Li G. Microarray-based functional gene analysis of soil microbial communities during ozonation and biodegradation of crude oil. Chemosphere, 2009, 75(2): 193–199
He Y, Xu J, Tang C, Wu Y. Facilitation of pentachlorophenol degradation in the rhizosphere of ryegrass (Lolium perenne L.). Soil Biology & Biochemistry, 2005, 37(11): 2017–2024
Liang R, Li C. Differences in cluster-root formation and carboxylate exudation in Lupinus albus L. under different nutrient deficiencies. Plant and Soil, 2003, 248(1–2): 221–227
Lu R. Soil Agricultural Chemical Analysis. Nanjing: China Agricultural Science and Technology Press, 1999 (in Chinese)
Coulon F, Pelletier E, Gourhant L, Delille D. Effects of nutrient and temperature on degradation of petroleum hydrocarbons in contaminated sub-Antarctic soil. Chemosphere, 2005, 58(10): 1439–1448
Zhou J, BrunsM A, Tiedje J M. DNA recovery from soils of diverse composition. Applied and Environmental Microbiology, 1996, 62(2): 316–322
Wu L, Liu X, Schadt C W, Zhou J. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Applied and Environmental Microbiology, 2006, 72(7): 4931–4941
Waldron P J, Wu L, van Nostrand J D, Schadt CW, He Z, Watson D B, Jardine P M, Palumbo A V, Hazen T C, Zhou J. Functional gene array-based analysis of microbial community structure in ground-waters with a gradient of contaminant levels. Environmental Science & Technology, 2009, 43(10): 3529–3534
Kirkpatrick W D, White P M Jr, Wolf D C, Thoma G J, Reynolds C M. Petroleum-degrading microbial numbers in rhizosphere and nonrhizosphere crude oil-contaminated soil. International Journal of Phytoremediation, 2008, 10(3): 210–221
Krutz L J, Beyrouty C A, Gentry T J, Wolf D C, Reynolds C M. Selective enrichment of a pyrene degrader population and enhanced pyrene degradation in Bermuda grass rhizosphere. Biology and Fertility of Soils, 2005, 41(5): 359–364
Whitman W B, Coleman D C, Wiebe W J. Prokaryotes: the unseen majority. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(12): 6578–6583
Li G, Huang W, Lerner D N, Zhang X. Enrichment of degrading microbes and bioremediation of petrochemical contaminants in polluted soil. Water Research, 2000, 34(15): 3845–3853
Ter Braak C J F. Canonical correspondence analysis: A new eigenvector technique for multivariate direct gradient analysis. Ecology, 1986, 67(5): 1167–1179
Liang Y, Li G, van Nostrand J D, He Z, Wu L, Deng Y, Zhang X, Zhou J. Microarray-based analysis of microbial functional diversity along an oil contamination gradient in oil field. FEMS Microbiology Ecology, 2009, 70(2): 324–333
van Hamme J D, Singh A, Ward O P. Recent advances in petroleum microbiology. Microbiology and Molecular Biology Reviews, 2003, 67(4): 503–549
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhong, Y., Wang, J., Song, Y. et al. Microbial community and functional genes in the rhizosphere of alfalfa in crude oil-contaminated soil. Front. Environ. Sci. Eng. 6, 797–805 (2012). https://doi.org/10.1007/s11783-012-0405-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11783-012-0405-z