Abstract
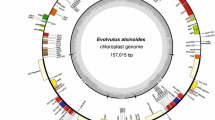
Carpesium abrotanoides L. is a widely used medicinal plant in China belonging to the tribe Inuleae of Family Asteraceae. Here, we determined the complete chloroplast (cp) genome sequence of C. abrotanoides and conducted a comparative analysis with closely related species from tribe Inuleae. The cp genome of C. abrotanoides exhibited a typical quadripartite structure with a full length of 151,394 bp, containing a total number of 133 genes. The gene order and GC content were highly consistent in the Inuleae plants according to the comparative analysis results. The number of potential RNA editing sites in 20 protein-coding genes of Inuleae species ranged from 43 in Inula hupehensis to 51 in Inula racemosa. In addition, 41 SSRs and 40 long repeats have been identified in the cp genomes of C. abrotanoides, exhibiting significant differences with that from other Inuleae plants. Six potential hotspots with high Pi values were identified among C. abrotanoides and other Inuleae species, including three intergenic spacer regions, two tRNA regions and one protein-coding region(ycf1). Furthermore, five pairs of primers targeting the divergence regions were accordingly designed, providing potential molecular markers for population genetic investigations in Inuleae species. The phylogenetic analysis based on conserved chloroplast protein coding genes revealed a sister relationship between C. abrotanoides and three Inula species with strong bootstrap support, indicating a close relationship between the two genera. This study enhanced the understanding of the evolution of Carpesium within the tribe Inuleae, identified several nucleotide diversity hotspots and provided the comparative information on the chloroplast genome, which could contribute to the molecular species discrimination and phylogenetic studies of Inuleae plants.






Similar content being viewed by others
Abbreviations
- LSC:
-
Large single copy
- SSC:
-
Small single copy
- IR:
-
Inverted repeat regions
- MAFFT:
-
Multiple Alignment using Fast Fourier Transform
- RSCU:
-
Relative synonymous codon usage
- Ks:
-
Synonymous substitution
- Ka:
-
Non-synonymous substitutions
- SNPs:
-
Single nucleotide polymorphisms
- InDels:
-
Insertions and deletions
- Ts:
-
Transitions substitutions
- Tv:
-
Transversion substitutions
- Ts/Tv:
-
Ratio of transition to transversion substitutions
- SSR:
-
Simple sequence repeats
References
Abdullah HCL, Mehmood F, Carlsen MM, Islam M, Waheed MT, Poczai P, Croat TB, Ahmed I (2020) Complete Chloroplast Genomes of Anthurium huixtlense and Pothos scandens (Pothoideae, Araceae): Unique Inverted Repeat Expansion and Contraction Affect Rate of Evolution. J Mol Evol 88:562–574. https://doi.org/10.1007/s00239-020-09958-w
Abdullah, Mehmood F, Rahim A, Heidari P, Ahmed I, Poczai P (2021) Comparative plastome analysis of Blumea, with implications for genome evolution and phylogeny of Asteroideae. Ecol Evol (12). https://doi.org/10.1002/ece3.7614
Abdullah WS, Mirza B, Ahmed I, Waheed MT (2019) Comparative analyses of chloroplast genomes of Theobroma cacao and Theobroma grandiflorum. Biologia 75:761–771. https://doi.org/10.2478/s11756-019-00388-8
Ajmal Ali M (2019) Molecular authentication of Anthemis deserti Boiss. (Asteraceae) based on ITS2 region of nrDNA gene sequence. Saudi J Biol Sci 26(1): 155–159. https://doi.org/10.1016/j.sjbs.2018.09.003
Amiryousefi A, Hyvönen J, Poczai P (2018) IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34:3030–3031. https://doi.org/10.1093/bioinformatics/bty220
Anderberg AA, Eldenäs P, Bayer RJ, Englund M (2004) Evolutionary relationships in the Asteraceae tribe Inuleae (incl. Plucheeae) evidenced by DNA sequences of ndh F; with notes on the systematic positions of some aberrant genera. Org Divers Evol 5(2): 135–146. https://doi.org/10.1016/j.ode.2004.10.015
Beier S, Thiel T, Münch T, Scholz U, Mascher M (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33:2583–2585. https://doi.org/10.1093/bioinformatics/btx198
Bi Y, Zhang M, Xue J, Dong R, Du Y, Zhang X (2018) Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci Rep 8:1184. https://doi.org/10.1038/s41598-018-19591-9
Chai X, Le Y, Wang J, Mei C, Feng J, Zhao H, Wang C, Lu D (2019) Carpesium abrotanoides (L.) Root as a Potential Source of Natural Anticancer Compounds: Targeting Glucose Metabolism and PKM2/HIF-1α Axis of Breast Cancer Cells. J Food Sci 84:3825–3832. https://doi.org/10.1111/1750-3841.14953
Chen Q, Wu X, Zhang D (2020) Comparison of the abilities of universal, super, and specific DNA barcodes to discriminate among the original species of Fritillariae cirrhosae bulbus and its adulterants. PLoS One 15(2). https://doi.org/10.1371/journal.pone.0229181
Chen X, Zhou J, Cui Y, Wang Y, Duan B, Yao H (2018) Identification of Ligularia Herbs Using the Complete Chloroplast Genome as a Super-Barcode. Front Pharmacol 9:695. https://doi.org/10.3389/fphar.2018.00695
Committee for the Pharmacopoeia of PR China (2015) Pharmacopoeia of PR China. Part I. People׳s Health Publishing House, PR, China, Beijing
Daniell H, Lin CS, Yu M, Chang WJ (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Bio 17:134. https://doi.org/10.1186/s13059-016-1004-2
Dastpak A, Osaloo SK, Maassoumi AA, Safar KN (2018) Molecular phylogeny of Astragalus sect, Ammodendron (Fabaceae) inferred from chloroplast ycf1 gene. Ann Bot Fenn 55:75–82. https://doi.org/10.5735/085.055.0108
Das A, Shakya A, Ghosh SK, Singh UP, Bhat HR (2020) A Review of Phytochemical and Pharmacological Studies of Inula Species. Curr Bioact Compd 16:557–567. https://doi.org/10.2174/1573407215666190207093538
Dong M, Zhou X, Ku W, Xu Z (2019) Detecting useful genetic markers and reconstructing the phylogeny of an important medicinal resource plant, Artemisia selengensis, based on chloroplast genomics. PLoS One 14:e0211340. https://doi.org/10.1371/journal.pone.0211340
Gao C, Ren X, Mason AS, Li J, Wang W, Xiao M, Fu D (2013) Revisiting an important component of plant genomes: microsatellites. Funct Plant Biol 40:645–661. https://doi.org/10.1071/FP12325
Gao C, Wang Q, Ying Z, Ge Y, Cheng R (2020) Molecular structure and phylogenetic analysis of complete chloroplast genomes of medicinal species Paeonia lactiflora from Zhejiang Province. Mitochondrial DNA B Resour 5:1077–1078. https://doi.org/10.1080/23802359.2020.1721372
Greiner S, Lehwark P, Bock R (2019) Organellar Genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res 47:W59–W64. https://doi.org/10.1093/nar/gkz238
Gutiérrez-Larruscain D, Santos-Vicente M, Anderberg AA, Rico E, Martínez-Ortega MM (2018) Phylogeny of the Inula group (Asteraceae: Inuleae): Evidence from nuclear and plastid genomes and arecircumscription of Pentanema. Taxon 67:149–164. https://doi.org/10.12705/671.9
He YQ, Cai L, Qian QG, Yang SH, Chen DL, Zhao BQ, Zhong ZP, Zhou XJ (2020) Anti-influenza A (H1N1) viral and cytotoxic sesquiterpenes from Carpesium abrotanoides. Phytochem Lett 35:41–45. https://doi.org/10.1016/j.phytol.2019.10.013
Henriquez CL, Abdullah AI, Carlsen MM, Zuluaga A, Croat TB, McKain MR (2020) Molecular evolution of chloroplast genomes in Monsteroideae (Araceae). Planta 251:72. https://doi.org/10.1007/s00425-020-03365-7
Huang S, Ge X, Cano A, Salazar BGM, Deng Y (2020) Comparative analysis of chloroplast genomes for five Diclipteraspecies (Acanthaceae): molecular structure, phylogenetic relationships, and adaptive evolution. PeerJ 8:e8450. https://doi.org/10.7717/peerj.8450
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. https://doi.org/10.1093/nar/gki198
Kearse MG, Wilusz JE (2017) Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31:1717–1731. https://doi.org/10.1101/gad.305250.117
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kurtz S (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642. https://doi.org/10.1093/nar/29.22.4633
Li B, Li Y, Cai Q, Lin F, Huang P, Zheng Y (2016) Development of chloroplast genomic resources for Akebia quinata (Lardizabalaceae). Conserv Genet Resour 8:447–449. https://doi.org/10.1007/s12686-016-0593-0
Li B, Lin F, Huang P, Guo W, Zheng Y (2017) Complete Chloroplast Genome Sequence of Decaisnea insignis: Genome Organization, Genomic Resources and Comparative Analysis. Sci Rep 7:10073. https://doi.org/10.1038/s41598-017-10409-8
Li C, Zheng Y, Huang P (2020a) Molecular markers from the chloroplast genome of rose provide a complementary tool for variety discrimination and profiling. Sci Rep 10:12188. https://doi.org/10.1038/s41598-020-68092-1
Li DM, Zhu GF, Xu YC, Ye YJ, Liu JM (2020b) Complete Chloroplast Genomes of Three Medicinal Alpinia Species: Genome Organization, Comparative Analyses and Phylogenetic Relationships in Family Zingiberaceae. Plants (basel) 9:286. https://doi.org/10.3390/plants9020286
Liang H, Zhang Y, Deng J, Gao G, Ding C, Zhang L, Yang R (2020) The Complete Chloroplast Genome Sequences of 14 Curcuma Species: Insights Into Genome Evolution and Phylogenetic Relationships Within Zingiberales. Front Genet 11:802. https://doi.org/10.3389/fgene.2020.00802
Liu H, Lu Y, Lan B, Xu J (2020) Codon usage by chloroplast gene is bias in Hemiptelea davidii. J Genet 99:8. https://doi.org/10.1007/s12041-019-1167-1
Loeuille B, Thode V, Siniscalchi C, Andrade S, Rossi M, Pirani JR (2021) Extremely low nucleotide diversity among thirty-six new chloroplast genome sequences from Aldama (Heliantheae, Asteraceae) and comparative chloroplast genomics analyses with closely related genera. PeerJ 9:e10886. https://doi.org/10.7717/peerj.10886
Mower JP (2009) The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res 37:W253–W259. https://doi.org/10.1093/nar/gkp337
Nguyen HQ, Nguyen TNL, Doan TN, Nguyen TTN, Phạm MH, Le TL, Sy DT, Chu HH, Chu HM (2021) Complete chloroplast genome of novel Adrinandra megaphylla Hu species: molecular structure, comparative and phylogenetic analysis. Sci Rep 11:11731. https://doi.org/10.1038/s41598-021-91071-z
Nie Z-L, Funk V, Sun H, Deng T, Meng Y, Wen J (2013) Molecular phylogeny of Anaphalis (Asteraceae, Gnaphalieae) with biogeographic implications in the Northern Hemisphere. J Plant Res 126:17–32. https://doi.org/10.1007/s10265-012-0506-6
Nie X, Deng P, Feng K, Liu P, Du X, You FM, Weining S (2014) Comparative analysis of codon usage patterns in chloroplast genomes of the Asteraceae family. Plant Mol Biol Report 32:828–840. https://doi.org/10.1007/s11105-013-0691-z
Nie L, Cui Y, Wu L, Zhou J, Xu Z, Li Y, Li X, Wang Y, Yao H (2019) Gene losses and variations in chloroplast genome of parasitic plant macrosolen and phylogenetic relationships within santalales. Int J Mol Sci 20:5812. https://doi.org/10.3390/ijms20225812
Nylinder S, Anderberg AA (2015) Phylogeny of the Inuleae (Asteraceae) with special emphasis on the Inuleae-Plucheinae. Taxon 64:110–130. https://doi.org/10.12705/641.22
Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Report 15:8–15. https://doi.org/10.1007/BF02772108
Pornpongrungrueng P, Borchsenius F, Englund M, Anderberg AA, Gustafsson MHG (2007) Phylogenetic relationships in Blumea (Asteraceae: Inuleae) as evidenced by molecular and morphological data. Plant Syst Evol 269:223–243. https://doi.org/10.1007/s00606-007-0581-7
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147. https://doi.org/10.1016/S0169-5347(00)02097-8
Qian QG, Gong LM, Yang SH, Zhao BQ, Cai J, Zhang ZJ, Wang YH, Zhou XJ (2020) Two new compounds from Carpesium abrotanoides. Phytochem Lett 40:5–9. https://doi.org/10.1016/j.phytol.2020.08.012
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Seca AML, Grigore A, Pinto DCGA, Silva AMS (2014) The genus Inula and their metabolites: From ethnopharmacological to medicinal uses. J Ethnopharmacol 154:286–310. https://doi.org/10.1016/j.jep.2014.04.010
Shao W, Huang S, Zhang Y, Jiang J, Li H (2021) Comparative transcriptomics provides a strategy for phylogenetic analysis and SSR marker development in Chaenomeles. Sci Rep 11:16441. https://doi.org/10.1038/s41598-021-95776-z
Shen J, Zhang X, Landis JB, Zhang H, Deng T, Sun H, Wang H (2020) Plastome evolution in Dolomiaea (Asteraceae, Cardueae) using phylogenomic and comparative analyses. Front Plant Sci 11:376. https://doi.org/10.3389/fpls.2020.00376
Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, Li X, Zhang B, Xu J, Chen S (2017) Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules 22:1330. https://doi.org/10.3390/molecules22081330
Sheng J, Zheng X, Wang J, Zeng X, Zhou F, Jin S, Hu Z, Diao Y (2017) Transcriptomics and proteomics reveal genetic and biological basis of superior biomass crop Miscanthus. Sci Rep 7:13777. https://doi.org/10.1038/s41598-017-14151-z
Singh RB, Mahenderakar MD, Jugran AK, Singh RK, Srivastava RK (2020) Assessing genetic diversity and population structure of sugarcane cultivars, progenitor species and genera using microsatellite (SSR) markers. Gene 753:144800. https://doi.org/10.1016/j.gene.2020.144800
Su Y, Huang L, Wang Z, Wang T (2018) Comparative chloroplast genomics between the invasive weed Mikania micrantha and its indigenous congener Mikania cordata structure variation, identification of highly divergent regions, divergence time estimation, and phylogenetic analysis. Mol Phylogenet Evol 126:181–195. https://doi.org/10.1016/j.ympev.2018.04.015
Sun J, Wang Y, Liu Y, Xu C, Yuan Q, Guo L, Huang L (2020) Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci Rep 10:11466. https://doi.org/10.1038/s41598-020-67943-1
Thode VA, Oliveira CT, Loeuille B, Siniscalchi CM, Pirani JR (2021) Comparative analyses of Mikania (Asteraceae: Eupatorieae) plastomes and impact of data partitioning and inference methods on phylogenetic relationships. Sci Rep 11:13267. https://doi.org/10.1038/s41598-021-92727-6
Wang L, Wuyun T, Du H, Wang D, Cao D (2016) Complete chloroplast genome sequences of Eucommia ulmoides: genome structure and evolution. Tree Genet Genomes 12:12. https://doi.org/10.1007/s11295-016-0970-6
Wang Z, Xu B, Li B, Zhou Q, Wang G, Jiang X, Wang C, Xu Z (2020) Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ 8:e8251. https://doi.org/10.7717/peerj.8251
Wu H, Wu H, Wang W, Liu T, Qi M, Feng J, Li X, Liu Y (2016) Insecticidal activity of sesquiterpene lactones and monoterpenoid from the fruits of Carpesium abrotanoides. Ind Crop Prod 92:77–83. https://doi.org/10.1016/j.indcrop.2016.07.046
Xu Y, Lu Y (2019) Noteworthy plants in Asteraceae from China. Journal of Zhejiang University(Science Edition) 46:200–205. https://doi.org/10.3785/j.issn.1008⁃9497.2019.02.008
Yan M, Fritsch PW, Moore MJ, Feng T, Meng A, Yang J, Deng T, Zhao C, Yao X, Sun H, Wang H (2018) Plastid phylogenomics resolves infrafamilial relationships of the Styracaceae and sheds light on the backbone relationships of the Ericales. Mol Phylogenet Evol 121:198–211. https://doi.org/10.1016/j.ympev.2018.01.004
Yang X, Luo X, Cai X (2014) Analysis of codon usage pattern in Taenia saginata based on a transcriptome dataset. Parasit Vectors 7:527–537. https://doi.org/10.1186/s13071-014-0527-1
Yang Y, Wang J, Li H, Mo Q, Huang H, Tao M, Luo Q, Liu H (2020) Five new C17/C15 sesquiterpene lactone dimers from Carpesium abrotanoides. Fitoterapia 145:104630. https://doi.org/10.1016/j.fitote.2020.104630
Yu X, Tan W, Zhang H, Gao H, Wang W, Tian X (2019) Complete chloroplast genomes of Ampelopsis humulifolia and Ampelopsis japonica: Molecular Structure, Comparative Analysis, and Phylogenetic Analysis. Plants 8(10):410. https://doi.org/10.3390/plants8100410
Zhang JP, Wang GW, Tian XH, Yang YX, Liu QX, Chen LP, Li HL, Zhang WD (2015) The genus Carpesium: A review of its ethnopharmacology, phytochemistry and pharmacology. J Ethnopharmacol 163:173–191. https://doi.org/10.1016/j.jep.2015.01.027
Zhang X, Deng T, Moore MJ, Ji Y, Lin N, Zhang H, Meng A, Wang H, Sun Y, Sun H (2019a) Plastome phylogenomics of Saussurea (Asteraceae: Cardueae). BMC Plant Biol 19:290. https://doi.org/10.1186/s12870-019-1896-6
Zhang Y, Yuan Y, Pang Y, Yu F, Yuan C, Wang D, Hu X (2019b) Phylogenetic Reconstruction and Divergence Time Estimation of Blumea DC. (Asteraceae: Inuleae) in China Based on nrDNA ITS and cpDNA trnL-F Sequences. Plants (Basel) 8:210. https://doi.org/10.3390/plants8070210
Zhao Y, Yin J, Guo H, Zhang Y, Xiao W, Sun C, Wu J, Qu X, Yu J, Wang X, Xiao J (2015) The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci 5:696. https://doi.org/10.3389/fpls.2014.00696
Zhou X, Liu M, Lu X, Sun S, Cheng Y, Ya H (2020) Genome survey sequencing and identification of genomic SSR markers for Rhododendron micranthum. Biosci Rep 40: BSR20200988. https://doi.org/10.1042/BSR20200988
Zhou T, Wang J, Jia Y, Li W, Xu F, Wang X (2018) Comparative Chloroplast Genome Analyses of Species in Gentiana section Cruciata (Gentianaceae) and the Development of Authentication Markers. Int J Mol Sci 19:1962. https://doi.org/10.3390/ijms19071962
Acknowledgements
We thank Shuisheng Yu and Yuqing Ge for their assistance and helpful suggestions during work. This work was financially supported by the Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Traditional Chinese Pharmacology) Zhejiang Chinese Medical University (No. ZYAOX2018033).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. XH, CG and RC designed the experiments and organized the manuscript. RC and QW collected the samples of Carpesium abrotanoides. XH, SD and MZ performed the experiments and conduced the comparative analysis. XH, SD and CG wrote the manuscript. RC was involved in writing-review and funding acquisition. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical statement
Ethics related study was not performed in current.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, X., Dong, S., Gao, C. et al. The complete chloroplast genome of Carpesium abrotanoides L. (Asteraceae): structural organization, comparative analysis, mutational hotspots and phylogenetic implications within the tribe Inuleae. Biologia 77, 1861–1876 (2022). https://doi.org/10.1007/s11756-022-01038-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-022-01038-2