Abstract
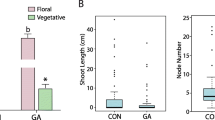
Brassinolide (BR) is crucial for regulating plant architecture. Apple dwarfing rootstocks are used to control apple tree size. However, information regarding the effects of BR on apple trees is limited. In addition, the molecular mechanism underlying the dwarfing of apple rootstocks is poorly understood. To elucidate the role of BR signal transduction genes in controlling apple tree architecture, five BR receptor kinase 1 (BRI1), nine BR-signaling kinase 1 (BSK1), two BRI1 KINASE INHIBITOR 1 (BKI1), and seven BR-insensitive 2 (BIN2) genes were analyzed. Bioinformatic analyses revealed that gene duplication events likely contributed to the expansion and evolution of the identified genes. Nine homologs between apple and Arabidopsis thaliana were also identified, and their expression patterns in different tissues were characterized. Exogenous BR treatments increased the primary shoot length and altered the expression of BR signal transduction genes (MdBRI1-5, MdBSK3-8, MdBKI1–2, MdBIN1–4, and MdBIN6/7). The scion of Fuji/Malling 9 (M.9) trees exhibited inhibited growth compared with that of Fuji/Fuji trees. The Fuji/M.9 trees had lower levels of the positive regulators of BR signaling (MdBRI1-5,MdBSK1, MdBSK4/7, and MdBSK6) and higher levels of the negative regulators (MdBIN5-7) compared with the Fuji/Fuji trees. Thus, the above-mentioned genes may help to regulate apple tree size in response to BR. In addition, MdBRI1–5, MdBSK1, MdBSK4/7, MdBSK6, and MdBIN5–7 have important roles in different grafting combinations. Our results may provide the basis for future analyses of BR signal transduction genes regarding their potential involvement in the regulation of plant architecture.

















Similar content being viewed by others
References
Cano-Delgado A et al (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131:5341–5351. doi:10.1242/dev.01403
Ceserani T, Trofka A, Gandotra N, Nelson T (2009) VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J 57:1000–1014. doi:10.1111/j.1365-313X.2008.03742.x
Clay NK, Nelson T (2002) VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell 14:2707–2722. doi:10.1105/tpc.005884
Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49:427–451. doi:10.1146/annurev.arplant.49.1.427
Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111:671–678
Colla G, Roupahel Y, Cardarelli M, Rea E (2006) Effect of salinity on yield, fruit quality, leaf gas exchange, and mineral composition of grafted watermelon plants. HortScience 41:622–627
Diaz-Hernandez B, Ciordia-Ara M, Coque-Fuertes M, Pereira-Lorenzo S (2003) Performance of six Asturian apple (Malus xdomestica) cultivars growing on two rootstocks for cider production. J Am Pomol Soc 57:121–127
Feng Y, Yin Y, Fei S (2015) Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci 234:163–173. doi:10.1016/j.plantsci.2015.02.015
Fujisawa H, Kudo K, Masuda T, Bessho H, Inomata Y (2010) Effects of JM1, JM 7, JM 8 and M.9 rootstocks on the photosynthesis rate of apple tree leaves. Hortic Res (Japan) 9:171–176
Hummer KE, Janick J (2009) Rosaceae: taxonomy, economic importance, genomics. In: Folta KM, Gardiner SE (eds) Plant genetics and genomics-crops and models, vol 6, pp 1–17. doi:10.1007/978-0-387-77491-6_1
Jaakola L, Pirttila AM, Halonen M, Hohtola A (2001) Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol 19:201–203. doi:10.1385/MB:19:2:201
Jeon H, Cho J, Park E, Han K, Choi Y, Ko J (2016) Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol J 14:1161–1170. doi:10.1111/pbi.12484
Jiang J et al (2015) The intrinsically disordered protein BKI1 is essential for inhibiting BRI1 signaling in plants. Mol Plant 8:1675–1678. doi:10.1016/j.molp.2015.07.012
Jin H, Do J, Shin SJ, Choi JW, Choi YI, Kim W, Kwon M (2014) Exogenously applied 24-epi brassinolide reduces lignification and alters cell wall carbohydrate biosynthesis in the secondary xylem of Liriodendron tulipifera. Phytochemistry 101:40–51. doi:10.1016/j.phytochem.2014.02.003
Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T (1996) Genetic evidence for an essential role of brassinosteroids in plant development. Plant J 9:701–713. doi:10.1046/j.1365-313X.1996.9050701.x
Kim TW et al (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11:1254–1260. doi:10.1038/ncb1970
Krost C, Petersen R, Schmidt ER (2012) The transcriptomes of columnar and standard type apple trees (Malus x domestica)—a comparative study. Gene 498:223–230. doi:10.1016/j.gene.2012.01.078
Lescot M et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295:1299–1301. doi:10.1126/science.1065769
Lyons E et al (2008) Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: coGe with rosids. Plant Physiol 148:1772–1781. doi:10.1104/pp.108.124867
Ma C et al (2014) Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (golden delicious). Mol Plant 7:218–230. doi:10.1093/mp/sst101
Ma Y, Xue H, Zhang L, Zhang F, Ou C, Wang F, Zhang Z (2016) Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus x domestica). Sci Rep 6:26719. doi:10.1038/srep26719
Manzano S, Martinez C, Megias Z, Gomez P, Garrido D, Jamilena M (2011) The role of ethylene and brassinosteroids in the control of sex expression and flower development in Cucurbita pepo. Plant Growth Regul 65:213–221. doi:10.1007/s10725-011-9589-7
Muhr M, Prufer N, Paulat M, Teichmann T (2016) Knockdown of strigolactone biosynthesis genes in populus affects BRANCHED1 expression and shoot architecture. New Phytol 212:613–626. doi:10.1111/nph.14076
Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2:1460–1471. doi:10.1371/journal.pbio.0020258
Noh SA, Choi YI, Cho JS, Lee H (2015) The poplar basic helix-loop-helix transcription factor BEE3—like gene affects biomass production by enhancing proliferation of xylem cells in poplar. Biochem Biophys Res Commun 462:64–70. doi:10.1016/j.bbrc.2015.04.109
Ohashi-Ito K, Kubo M, Demura T, Fukuda H (2005) Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol 46:1646–1656. doi:10.1093/pcp/pci180
Peng P, Yan Z, Zhu Y, Li J (2008) Regulation of the Arabidopsis GSK3-like kinase brassinosteroid-insensitive 2 through proteasome-mediated protein degradation. Mol Plant 1:338–346. doi:10.1093/mp/ssn001
Peng S, Tao P, Xu F, Wu A, Huo W, Wang J (2016) Functional characterization of soybean Glyma04g39610 as a brassinosteroid receptor gene and evolutionary analysis of soybean brassinosteroid receptors. Int J Mol Sci. doi:10.3390/ijms17060897
Pereira-Netto AB, Cruz-Silva C, Schaefer S, Ramirez JA, Galagovsky LR (2006) Brassinosteroid-stimulated branch elongation in the marubakaido apple rootstock. Trees-Struct Funct 20:286–291. doi:10.1007/s00468-005-0041-3
Pereira-Netto AB, Roessner U, Fujioka S, Bacic A, Asami T, Yoshida S, Clouse SD (2009) Shooting control by brassinosteroids: metabolomic analysis and effect of brassinazole on Malus prunifolia, the Marubakaido apple rootstock. Tree Physiol 29:607–620. doi:10.1093/treephys/tpn052
Santos A, Ribeiro R, Crespi AL (2004) Sweet cherry (Prunus avium) growth is mostly affected by rootstock and much less by budding height. New Zeal J Crop Hort 32:309–318
Shahnejat-Bushehri S, Tarkowska D, Sakuraba Y, Balazadeh S (2016) Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat Plants 2:16013. doi:10.1038/nplants.2016.13
Song C et al (2016) Expression analysis of key auxin synthesis, transport, and metabolism genes in different young dwarfing apple trees. Acta Physiol Plant. doi:10.1007/s11738-016-2065-2
Song C et al (2017) miRNA and degradome sequencing reveal miRNA and their target genes that may mediate shoot growth in spur type mutant “Yanfu 6”. Front Plant Sci. doi:10.3389/fpls.2017.00441
Sreeramulu S et al (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J 74:905–919. doi:10.1111/tpj.12175
Sun Y et al (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19:765–777. doi:10.1016/j.devcel.2010.10.010
Tan Y, Wang S, Liang D, Li M, Ma F (2014) Genome-wide identification and expression profiling of the cystatin gene family in apple (Malus x domestica Borkh.). Plant Physiol Bioch 79:88–97. doi:10.1016/j.plaphy.2014.03.011
Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH (2008a) Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 18:1944–1954. doi:10.1101/gr.080978.108
Tang W et al (2008b) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321:557–560. doi:10.1126/science.1156973
Ten Hove CA et al (2011) Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol Biol 76:69–83. doi:10.1007/s11103-011-9769-x
Velasco R et al (2010) The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet 42:833–839. doi:10.1038/ng.654
Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313:1118–1122. doi:10.1126/science.1127593
Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410:380–383. doi:10.1038/35066597
Wang L et al (2010) Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol 10:282. doi:10.1186/1471-2229-10-282
Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X (2013) Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 27:681–688. doi:10.1016/j.devcel.2013.11.010
Webster AD (1995) Rootstock and interstock effects on deciduous fruit tree vigour, precocity, and yield productivity. New Zeal J Crop Hort 23:373–382
Wei F et al (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3:e123. doi:10.1371/journal.pgen.0030123
Wei W, Hu Y, Han Y, Zhang K, Zhao F, Feng J (2016) The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: identification and expression analysis under biotic and abiotic stresses. Plant Physiol Bioch 105:129–144. doi:10.1016/j.plaphy.2016.04.014
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci USA 109:1187–1192. doi:10.1073/pnas.1109047109
Yang Z et al (2014) PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol 203:437–448. doi:10.1111/nph.12824
Yu X et al (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65:634–646. doi:10.1111/j.1365-313X.2010.04449.x
Zhang Y et al (2012) Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS One 7:e32153. doi:10.1371/journal.pone.0032153
Acknowledgements
Xilong Chen assisted with the analyses. This study was sponsored by the National Science and Technology Supporting Project (2013BAD20B03), the National Apple Industry Technology System of the Ministry of Agriculture of China (CARS-28), the National Spark Plan Program (2014GA850002), the Science and Technology Innovative Engineering Project in Shaanxi Province of China (2015NY114), the Yangling Subsidiary Center Project of the National Apple Improvement Center, the Collaborative Innovation Center of Shaanxi Fruit Industry Development, and the Innovation Project of Science and Technology of Shaanxi Province (2016TZC-N-11-6).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Chandra.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11738_2017_2479_MOESM1_ESM.docx
Table S1. Sequences of the primers used to amplify apple BR signal transduction genes in qRT-PCR analyses. The EF-1α gene was used as an internal control to normalize the BR signal transduction gene expression levels. Table S2. Best hits for each putative apple BR signal transduction gene, using a local search against apple expressed sequence taga assemblies. a Downloaded from NCBI. Table S3. Motif sequences identified using the MEME program. Conserved motifs identified in BR signal transduction proteins. Motif numbers correspond to the motifs in Fig. 2. Table S4. Analysis of the synteny among the apple BR signal transduction genes. Synteny blocks of BR signal transduction genes within the apple genome. Table S5. Analysis of the synteny among the BR signal transduction genes of apple and A. thaliana. Synteny blocks of BR signal transduction genes between the genomes of apple and A. thaliana. (DOCX 23 kb)
11738_2017_2479_MOESM2_ESM.tif
Fig. S1. Brassinolide signal transduction pathway. Filled objects represent the BR-signaling components. The gray circle and square represent the BZR1/2-binding element. Arrows indicate positive regulation, while the T-shaped lines indicate negative regulation. (TIFF 63 kb)
11738_2017_2479_MOESM3_ESM.tif
Fig. S2. Interaction network for proteins encoded by apple BR signal transduction genes based on A. thaliana orthologs. Line thickness is positively correlated with the value of the combined score. Homologous genes in apple are indicated in black text written in parentheses. (TIFF 19244 kb)
11738_2017_2479_MOESM4_ESM.tif
Fig. S3. Predicted cis-elements in apple BR signal transduction promoter sequences. The promoter sequences (−1500 bp upstream from the transcriptional start site) of the 23 identified BR signal transduction genes were analyzed. The names of the BR signal transduction genes are provided on the left side of the figure. The numbers at the bottom refer to the number of nucleotides upstream of the translation initiation codon, ATG. The hormone response elements are indicated by different colored shapes, similar to Figure S3. (TIFF 4711 kb)
Rights and permissions
About this article
Cite this article
Zheng, L., Ma, J., Song, C. et al. Genome-wide identification and expression profiling analysis of brassinolide signal transduction genes regulating apple tree architecture. Acta Physiol Plant 39, 177 (2017). https://doi.org/10.1007/s11738-017-2479-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2479-5